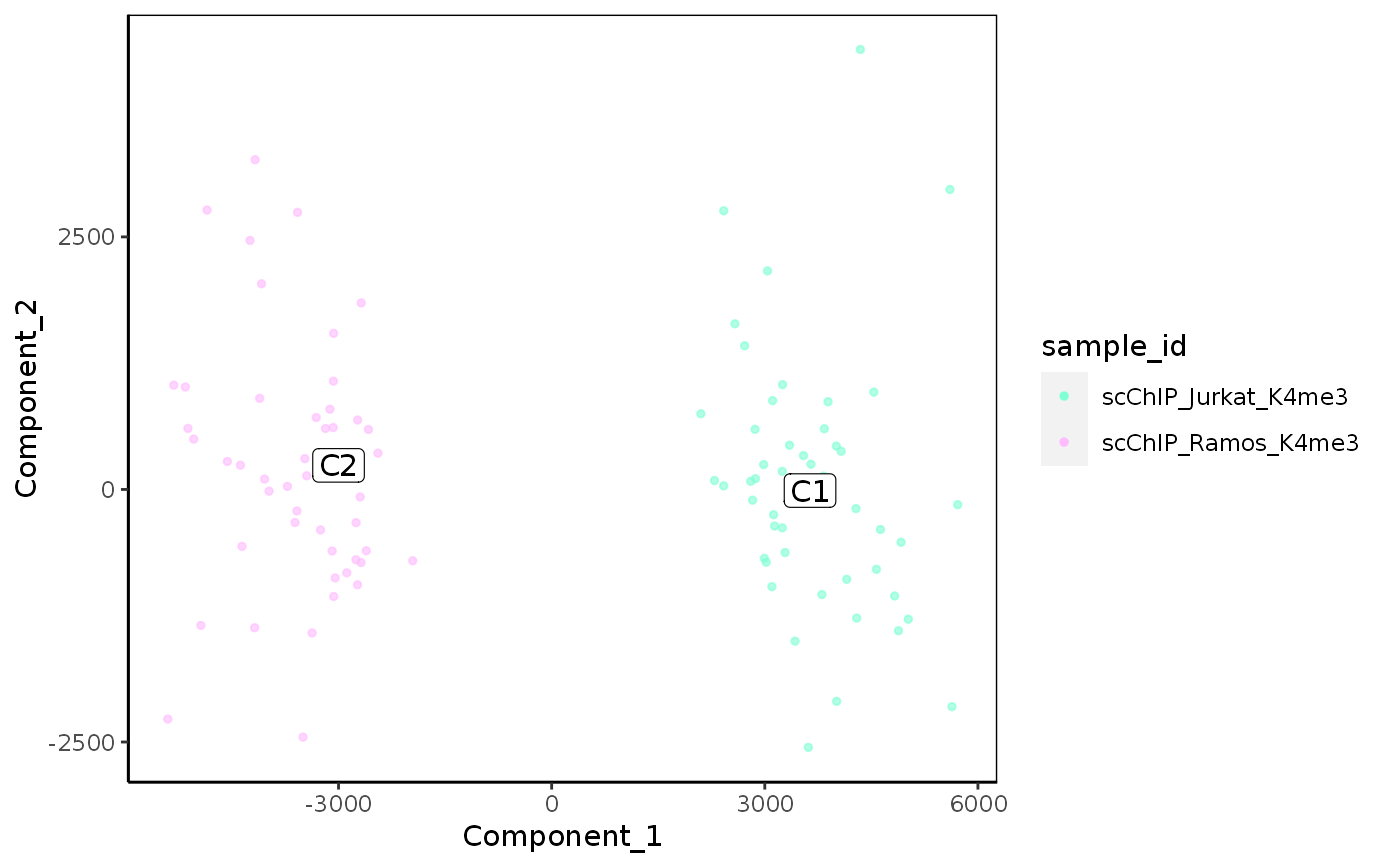
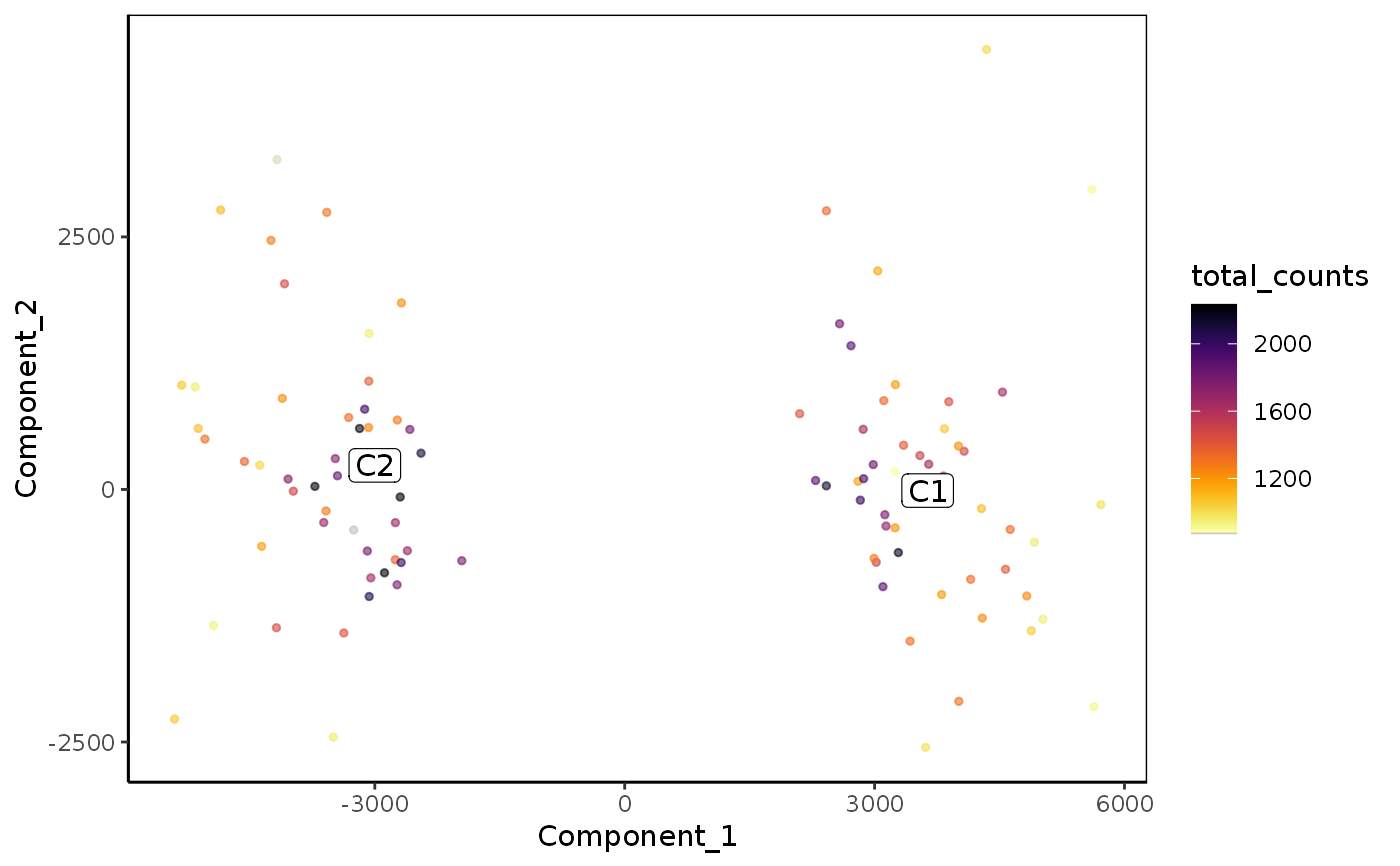
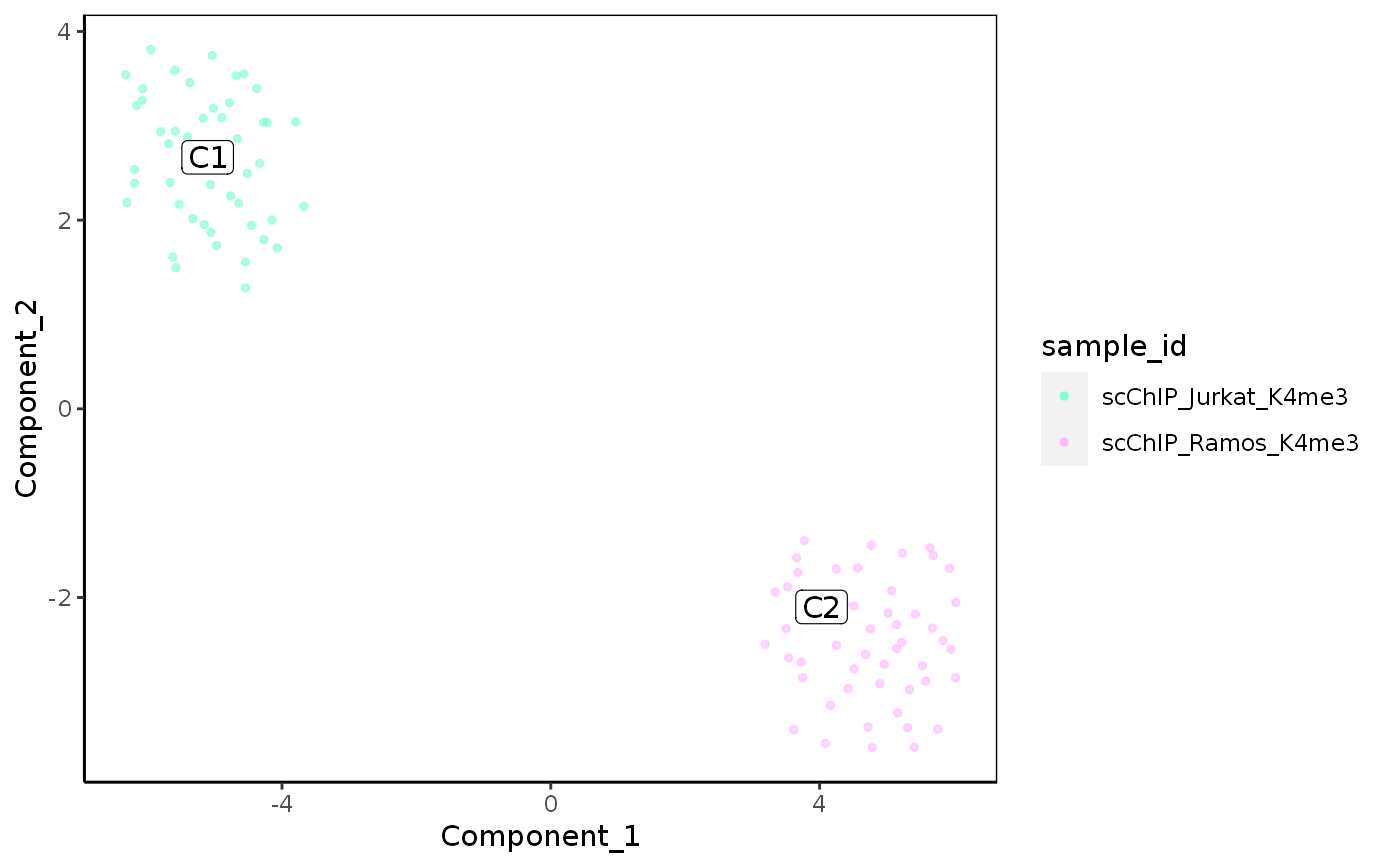
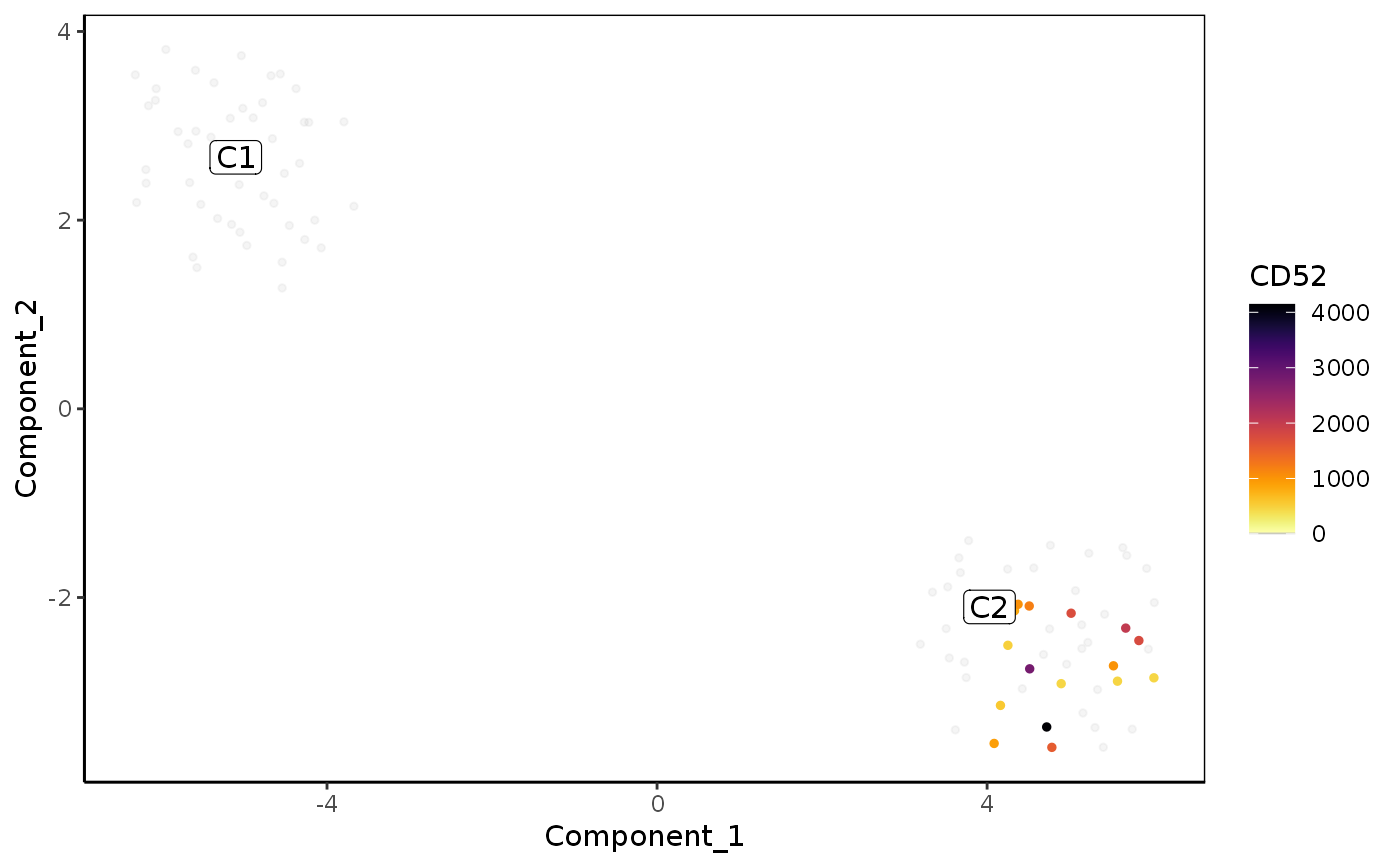
Plot reduced dimensions (PCA, TSNE, UMAP)
plot_reduced_dim_scExp( scExp, color_by = "sample_id", reduced_dim = c("PCA", "TSNE", "UMAP"), select_x = NULL, select_y = NULL, downsample = 5000, transparency = 0.6, size = 1, max_distanceToTSS = 1000, annotate_clusters = "cell_cluster" %in% colnames(colData(scExp)), min_quantile = 0.01, max_quantile = 0.99 )
Arguments
| scExp | A SingleCellExperiment Object |
|---|---|
| color_by | Character of eature used for coloration. Can be cell metadata ('total_counts', 'sample_id', ...) or a gene name. |
| reduced_dim | Reduced Dimension used for plotting |
| select_x | Which variable to select for x axis |
| select_y | Which variable to select for y axis |
| downsample | Number of cells to downsample |
| transparency | Alpha parameter, between 0 and 1 |
| size | Size of the points. |
| max_distanceToTSS | The maximum distance to TSS to consider a gene linked to a region. Used only if "color_by" is a gene name. |
| annotate_clusters | A logical indicating if clusters should be labelled. The 'cell_cluster' column should be present in metadata. |
| min_quantile | The lower threshold to remove outlier cells, as quantile of cell embeddings (between 0 and 0.5). |
| max_quantile | The upper threshold to remove outlier cells, as quantile of cell embeddings (between 0.5 and 1). |
Value
A ggplot geom_point plot of reduced dimension 2D reprensentation
Examples
plot_reduced_dim_scExp(scExp, color_by = "total_counts")plot_reduced_dim_scExp(scExp, reduced_dim = "UMAP")plot_reduced_dim_scExp(scExp, color_by = "CD52", reduced_dim = "UMAP")