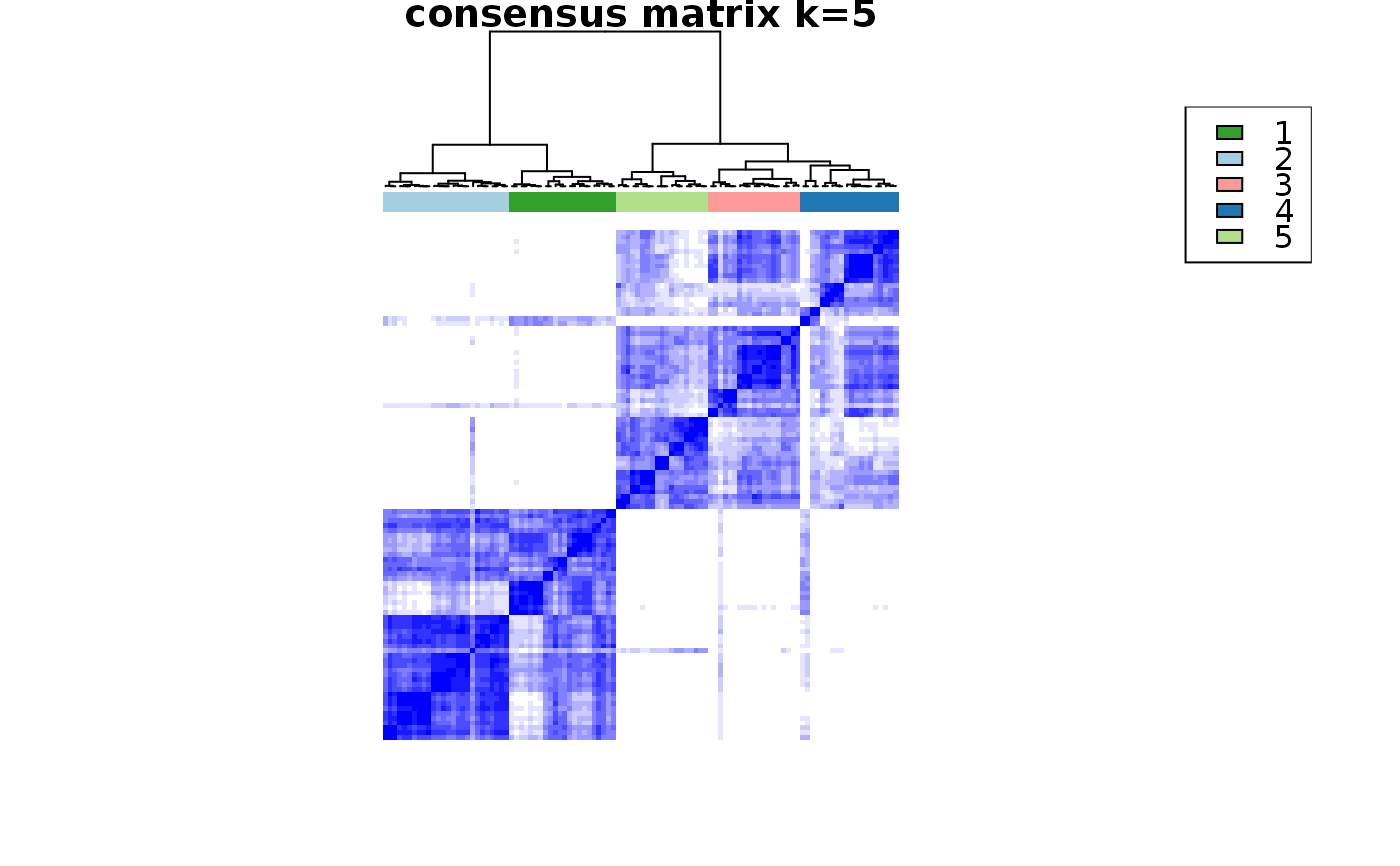
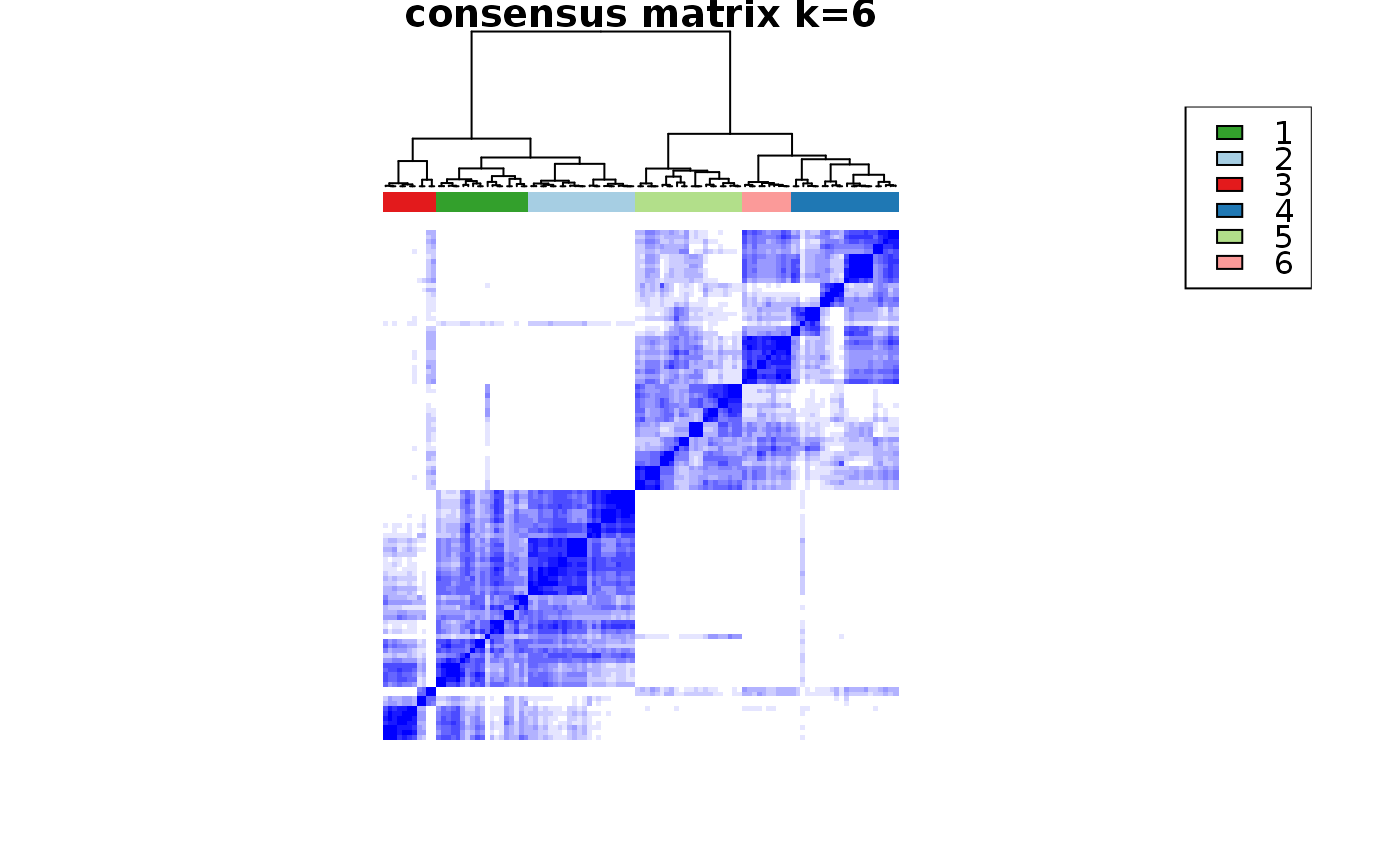
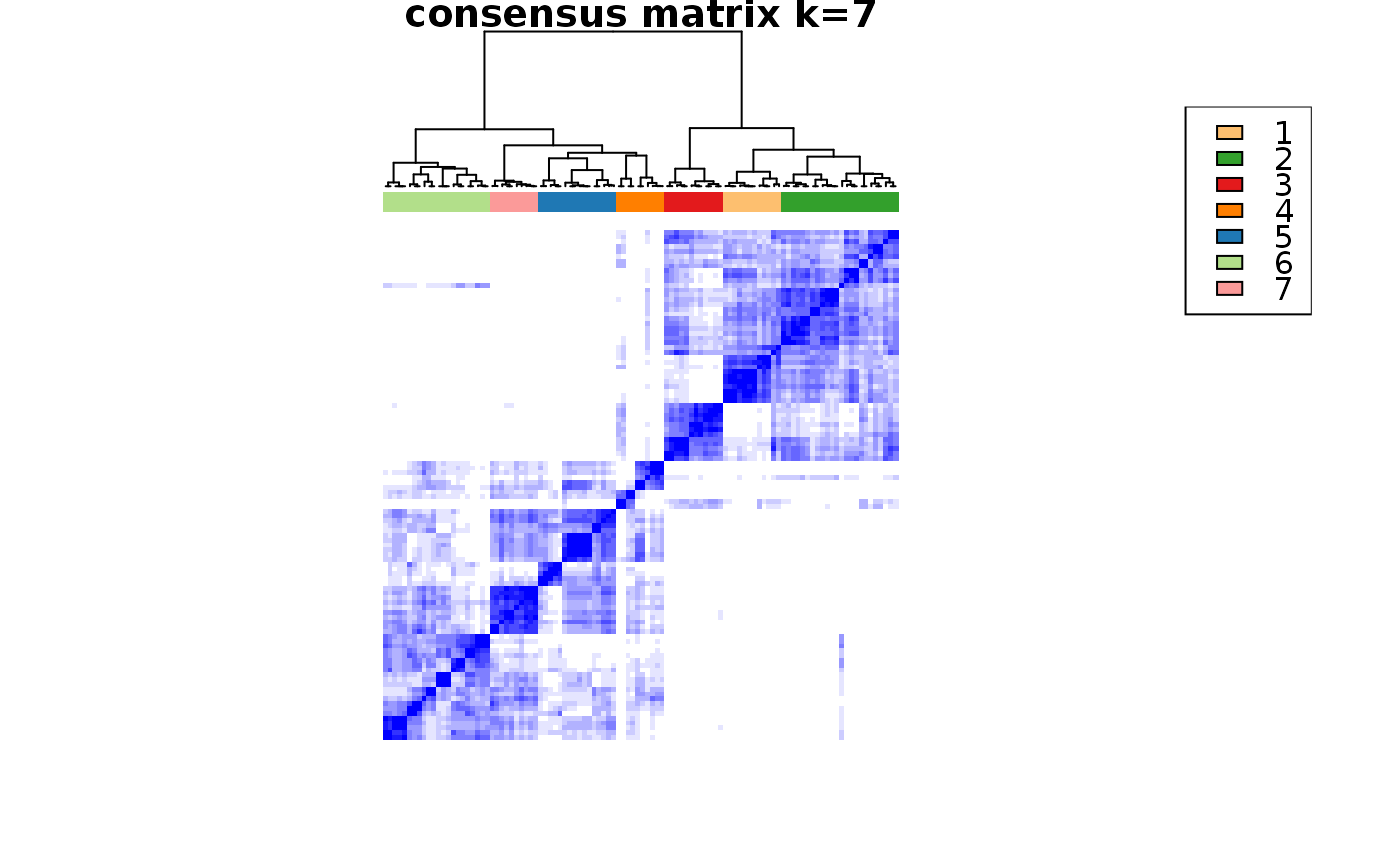
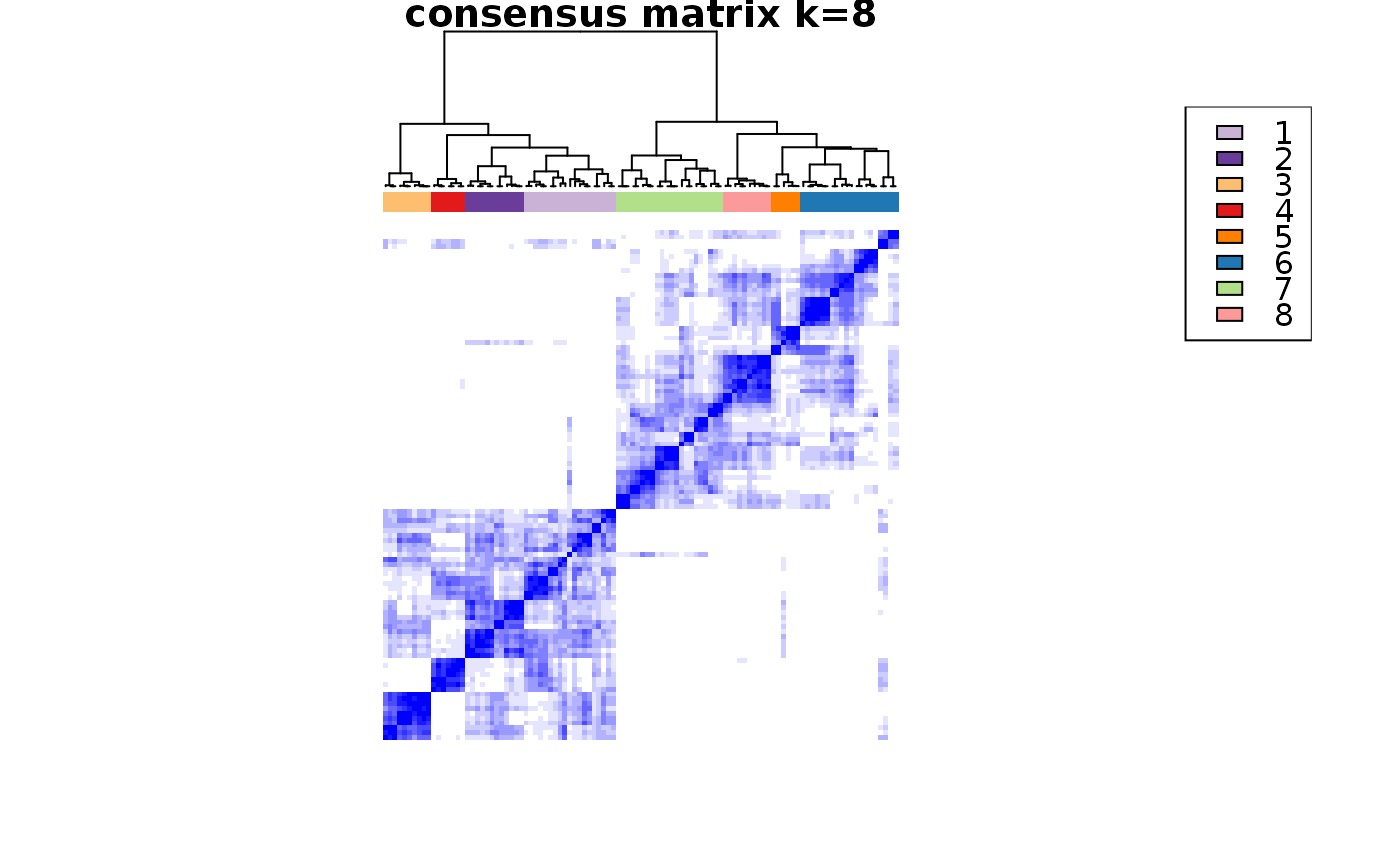
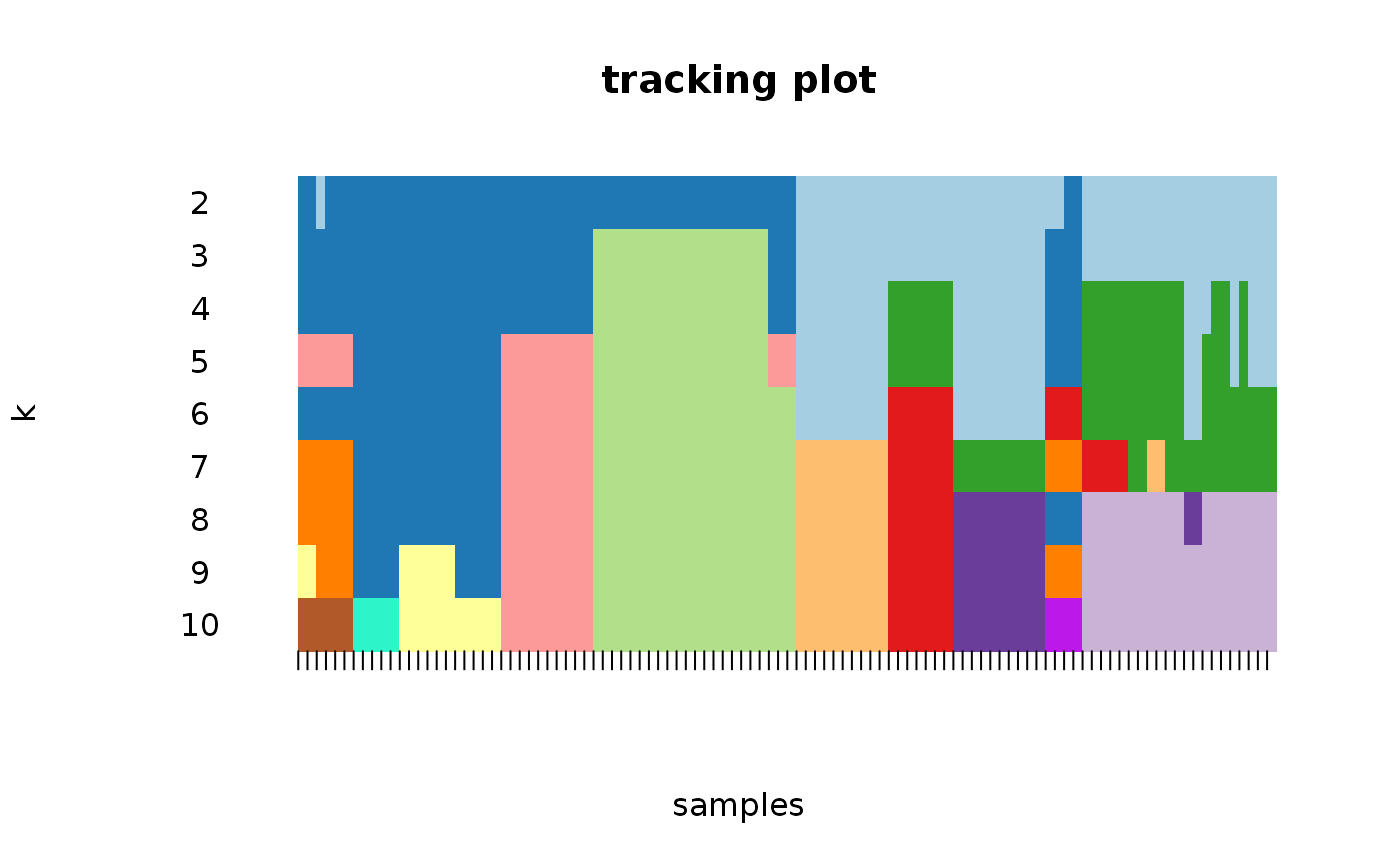
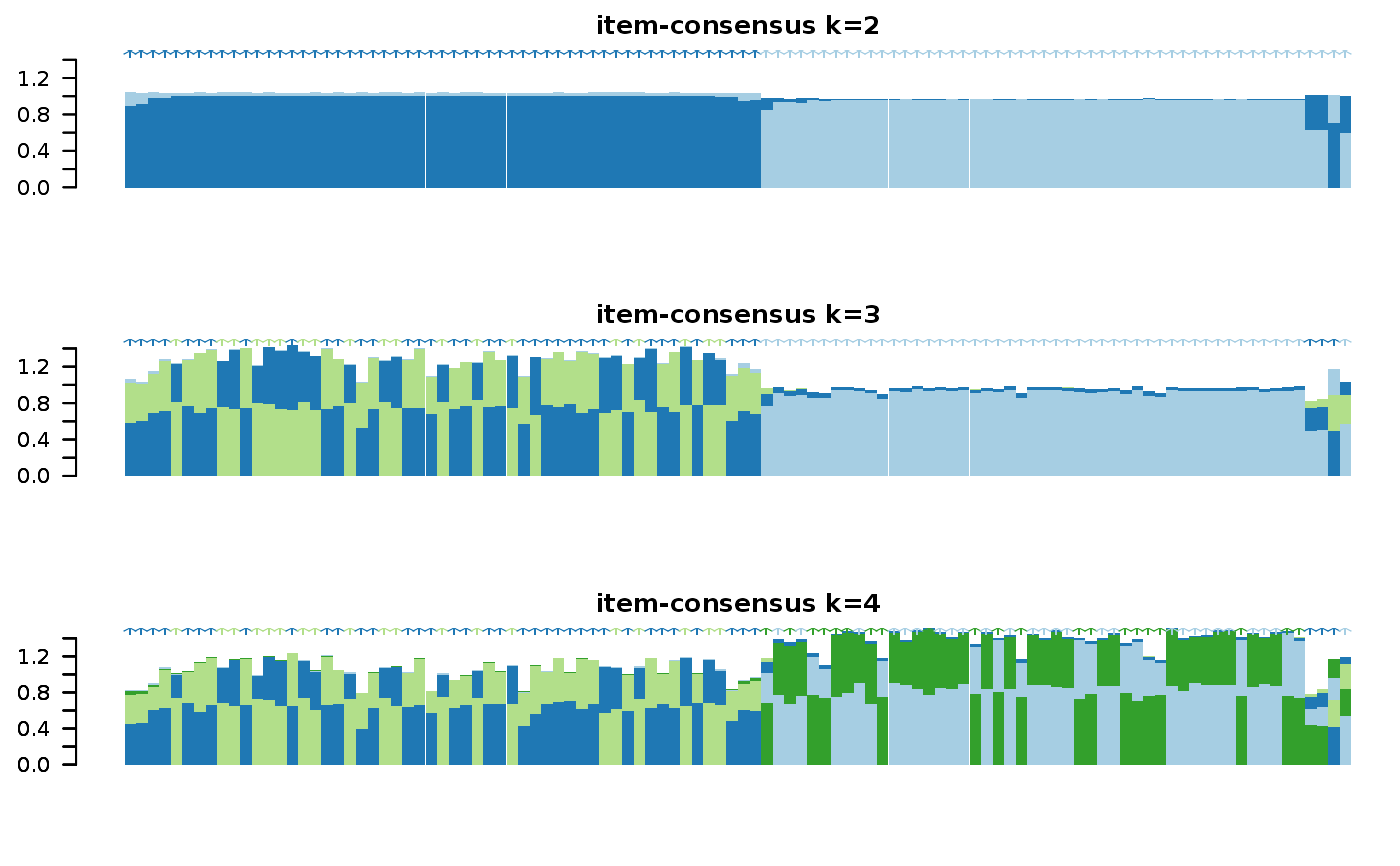
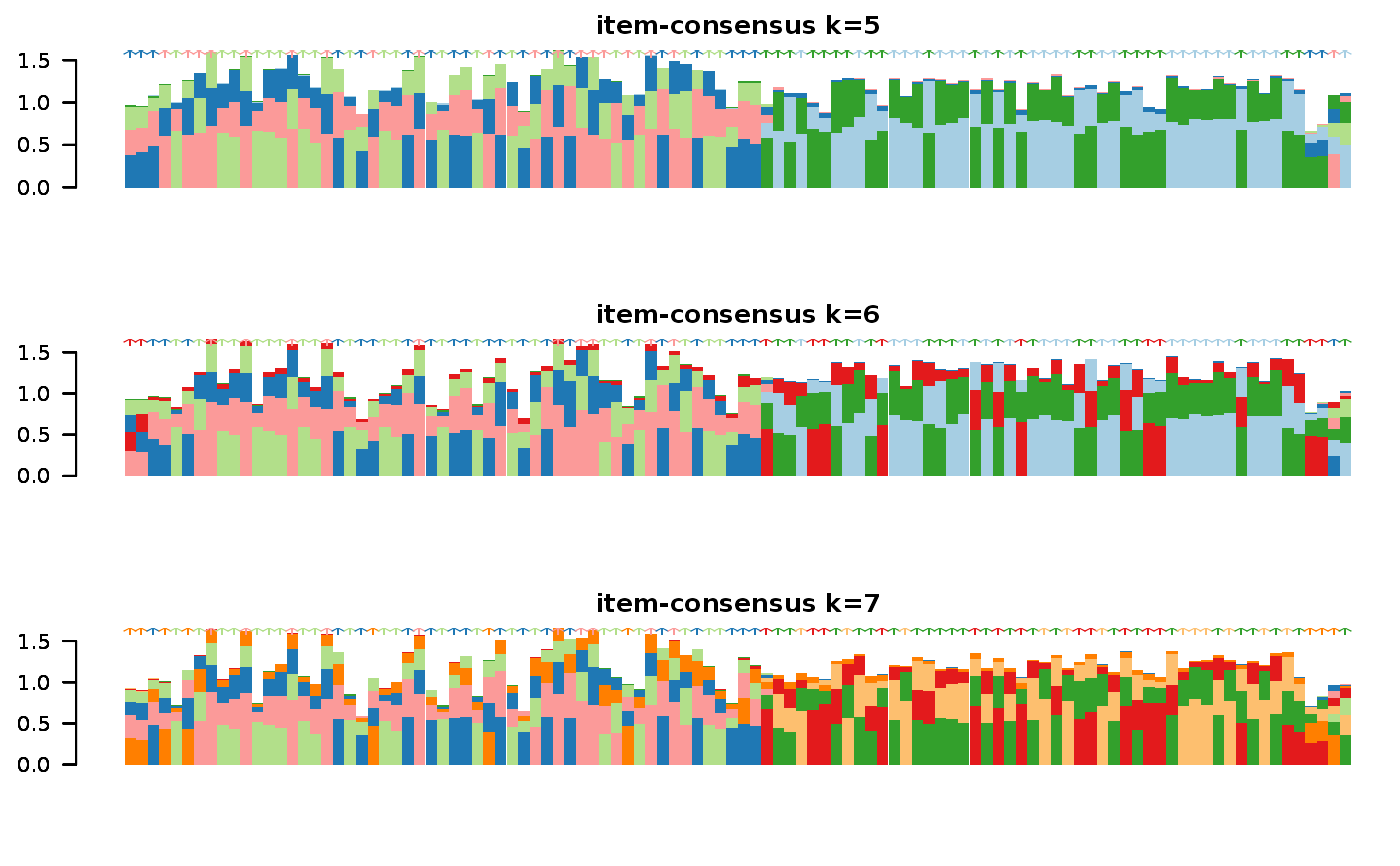
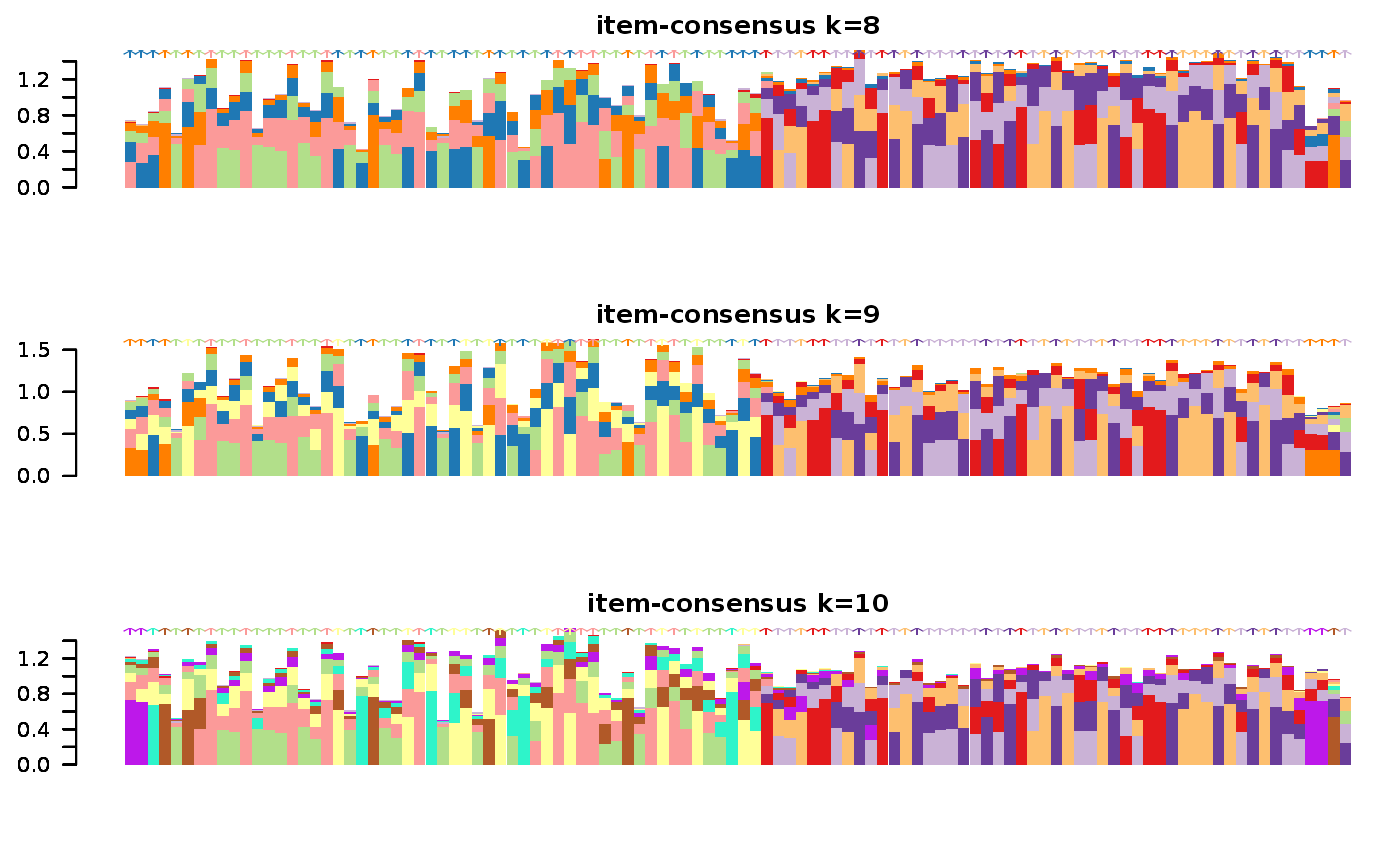
Wrapper to apply ConsensusClusterPlus to scExp object
Source:R/correlation_filtering_clustering.R
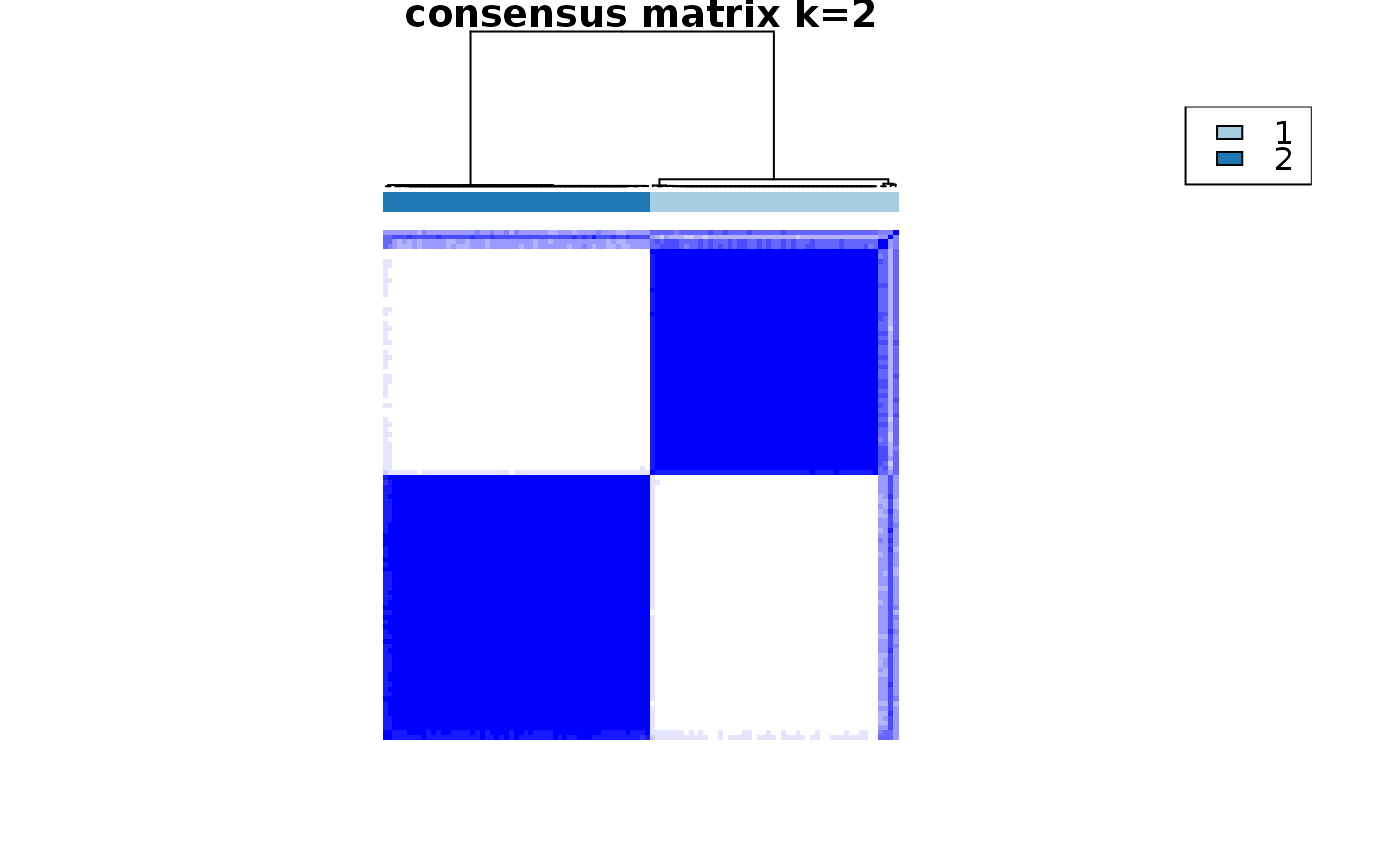
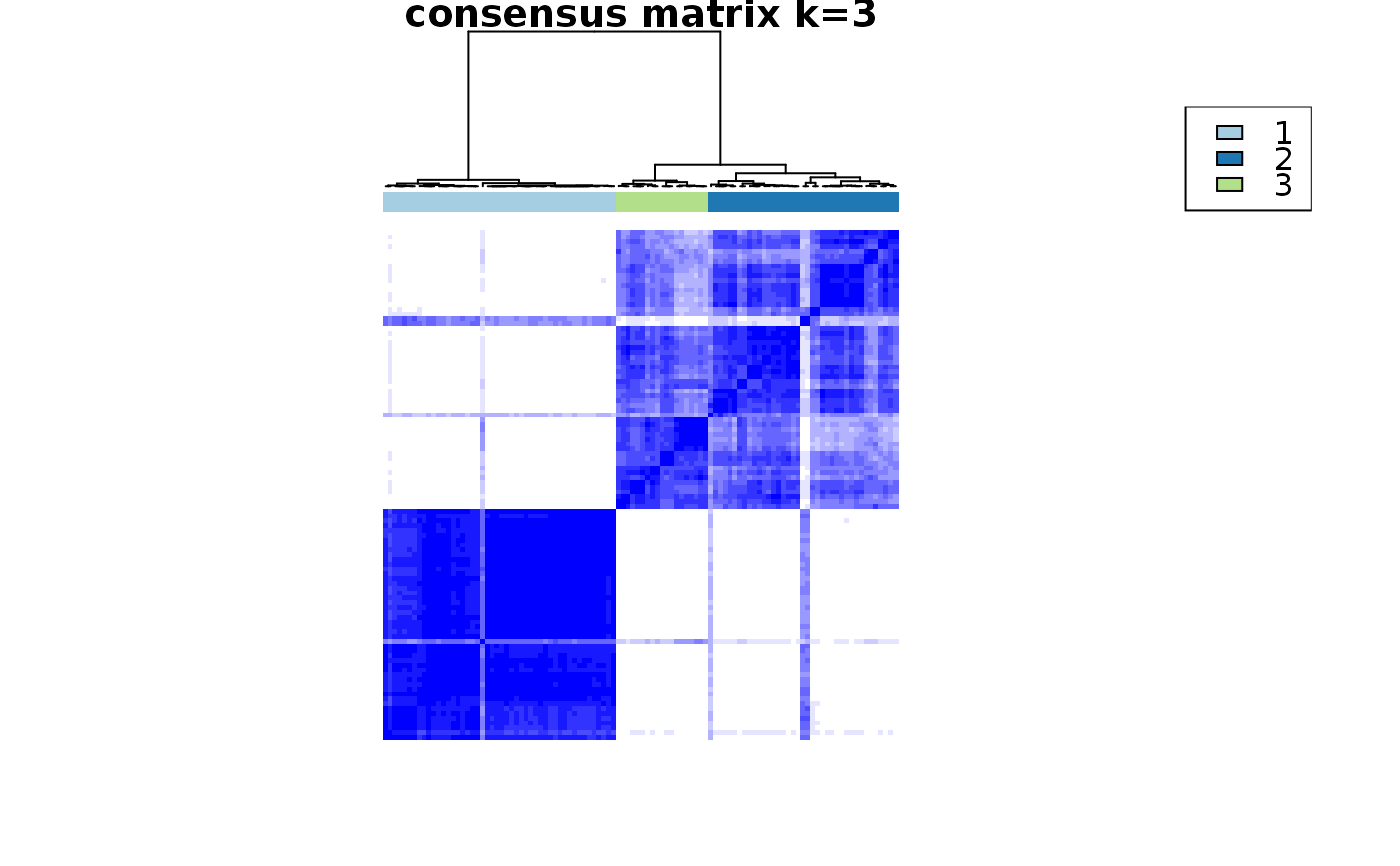
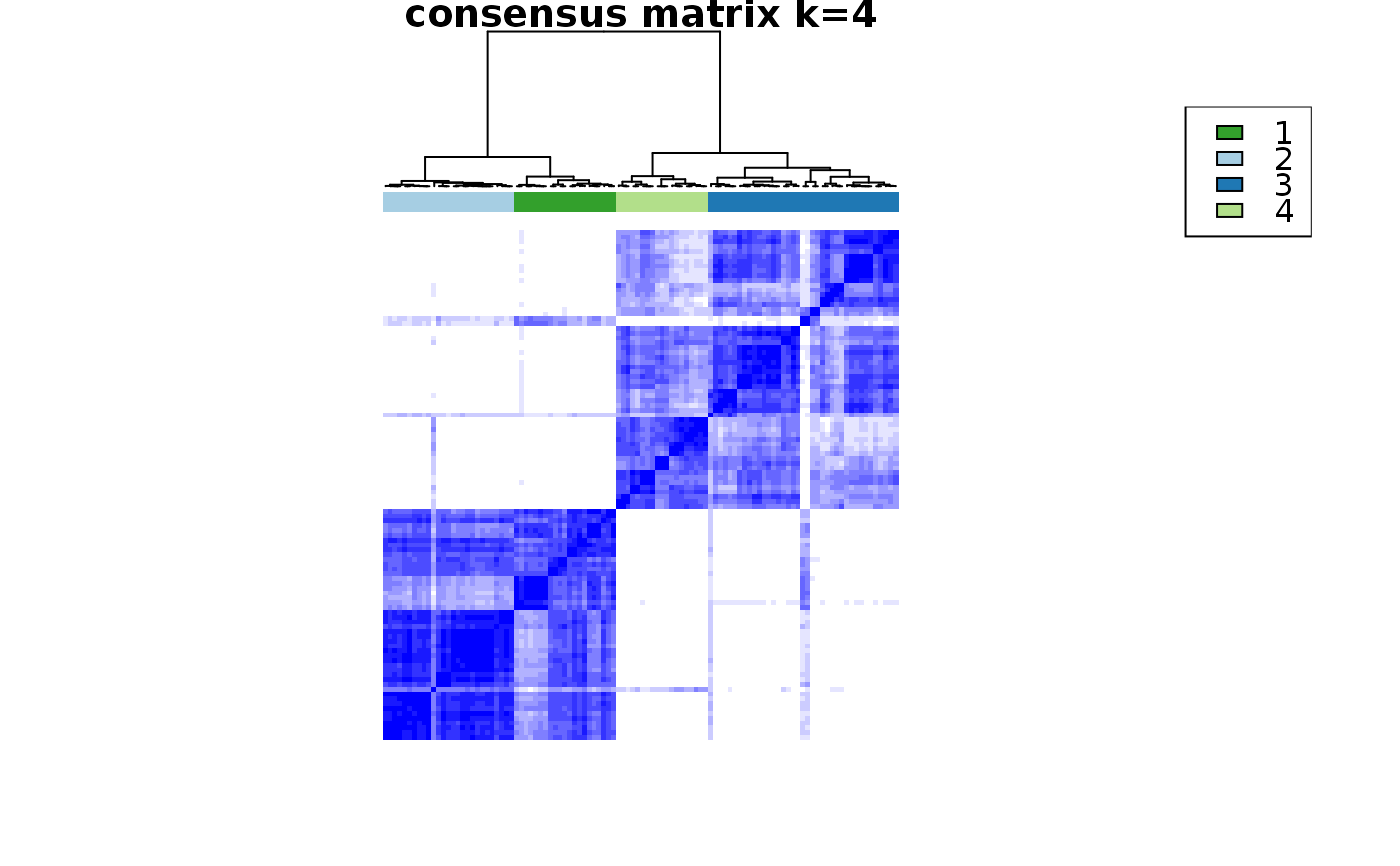
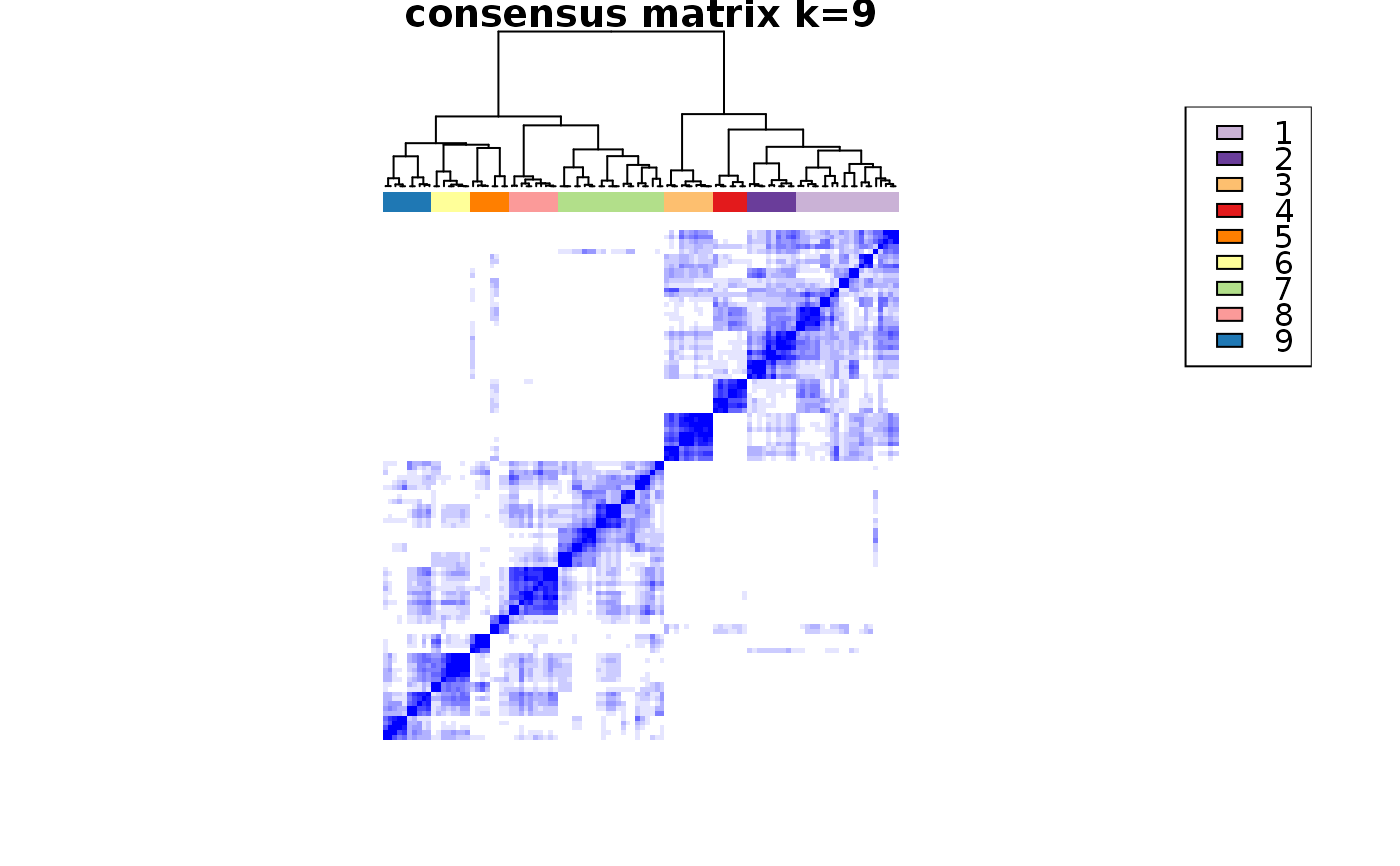
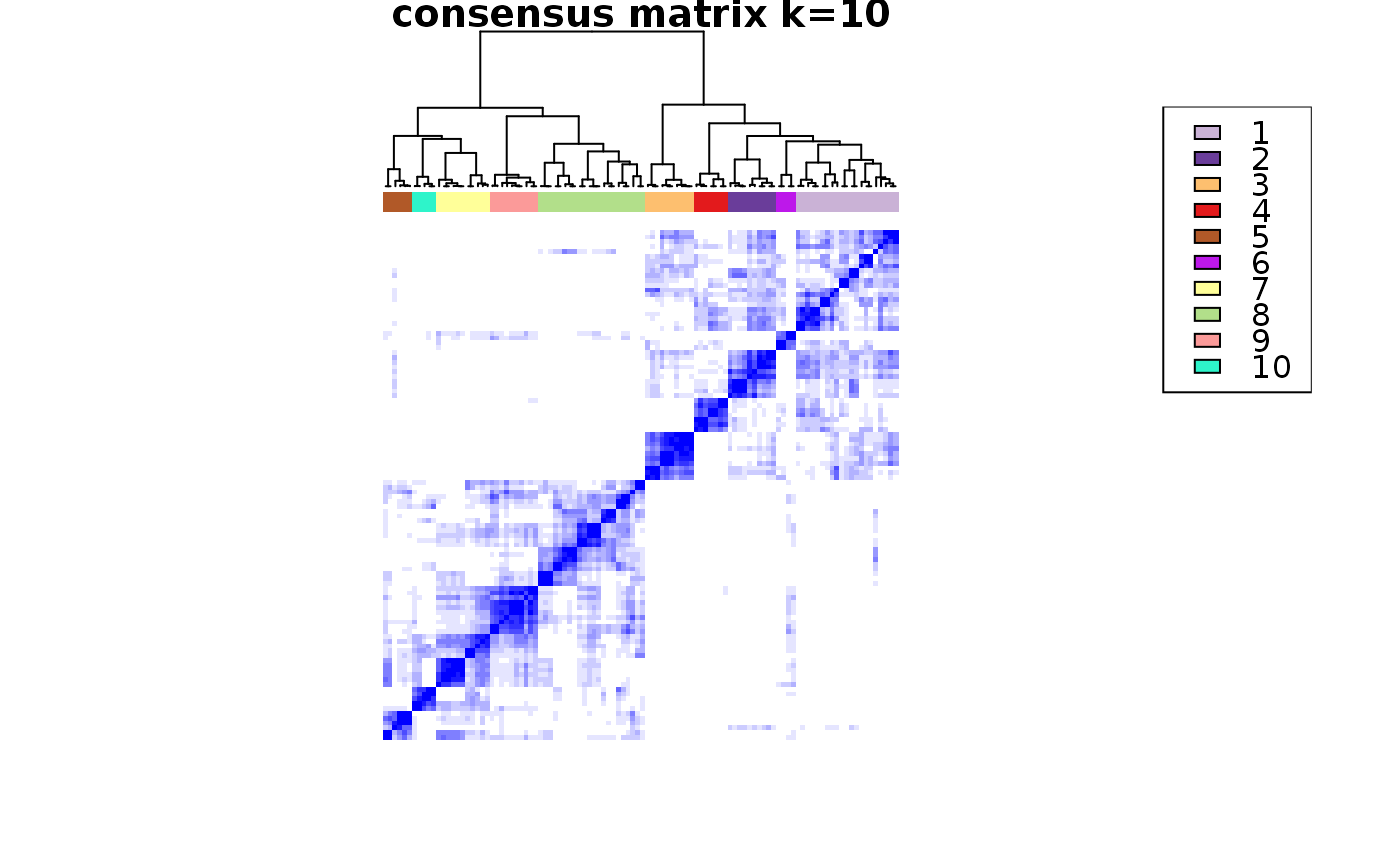
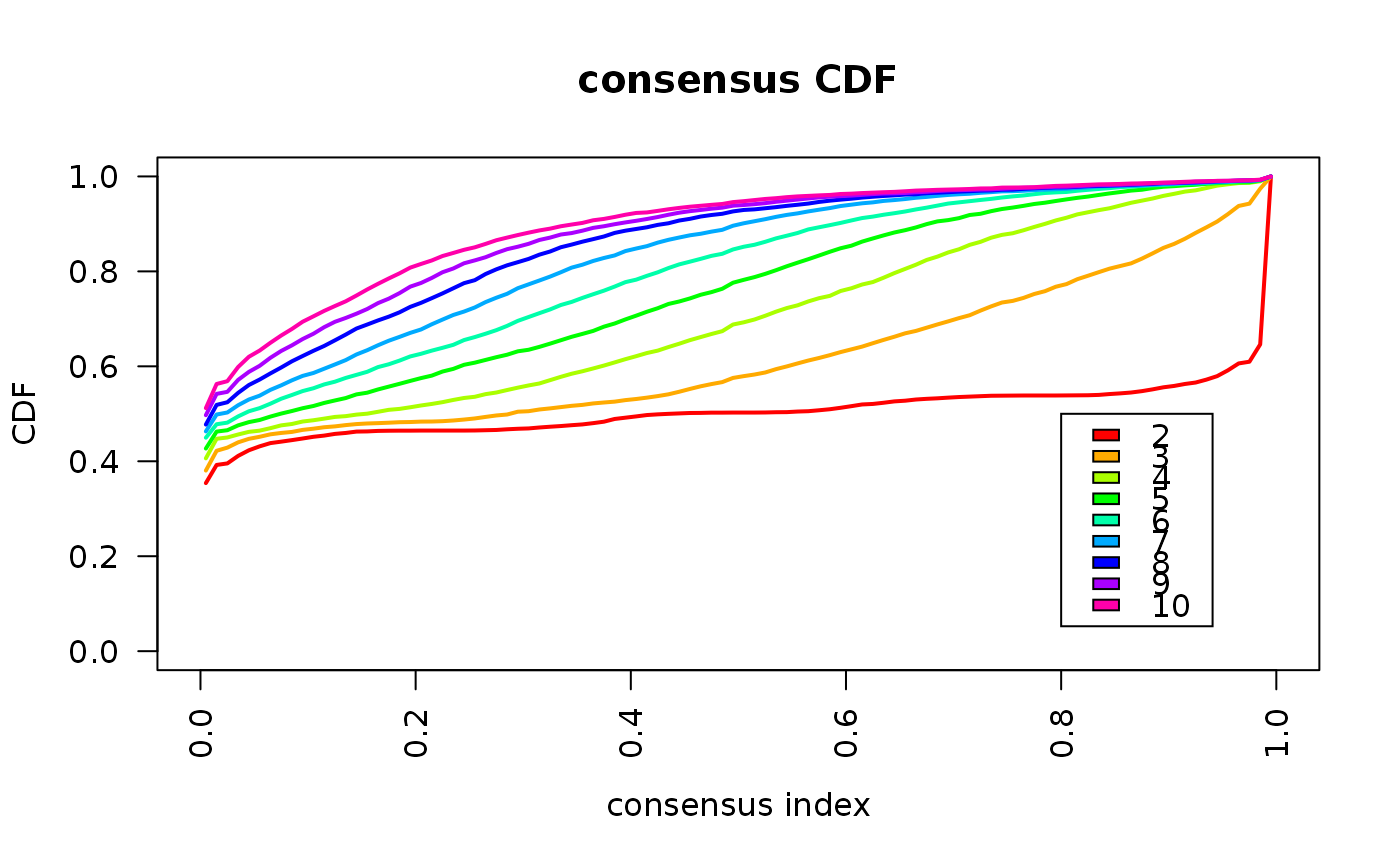
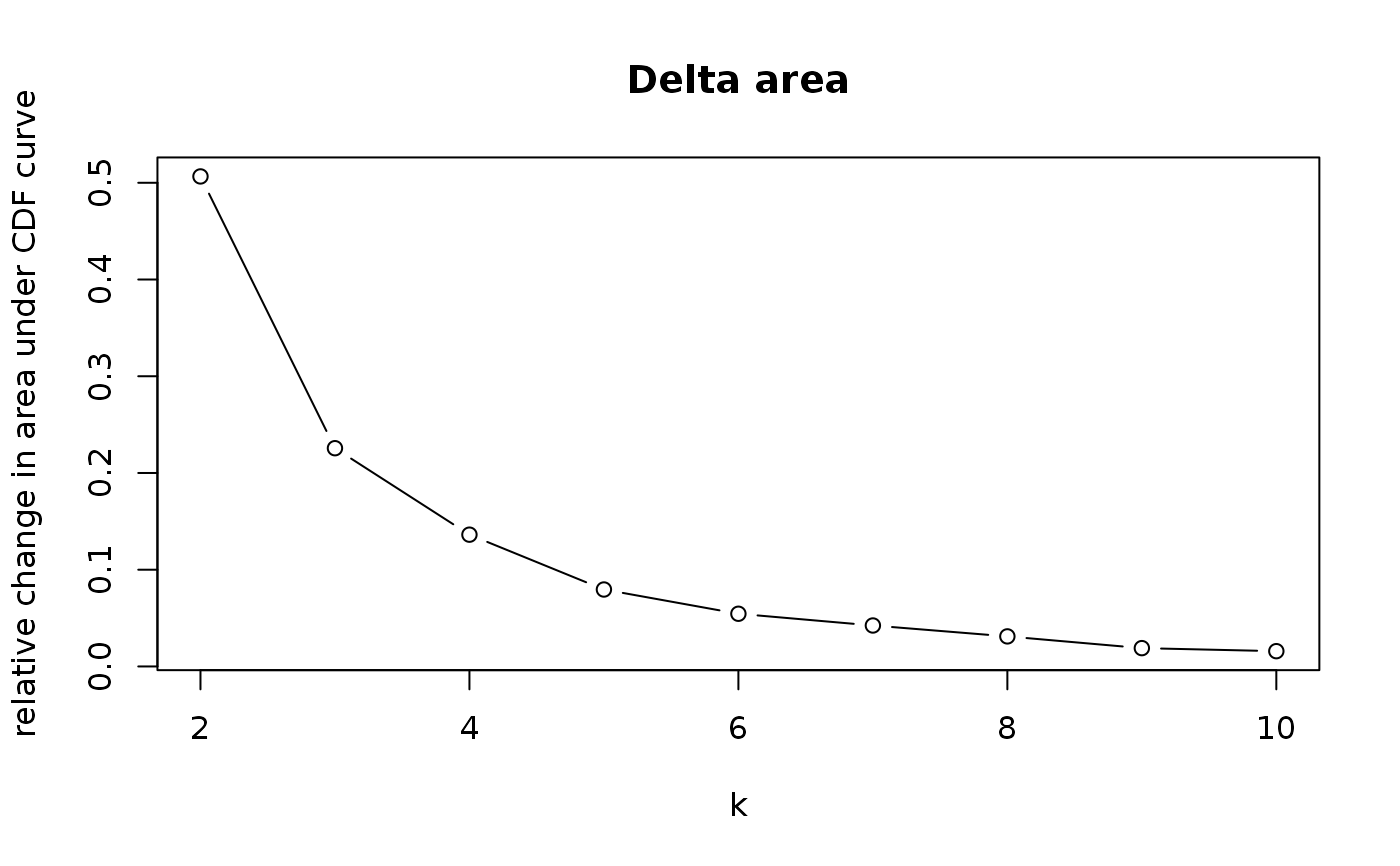
consensus_clustering_scExp.RdRuns consensus hierarchical clustering on PCA feature space of scExp object. Plot consensus scores for each number of clusters. See ConsensusClusterPlus - Wilkerson, M.D., Hayes, D.N. (2010). ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics, 2010 Jun 15;26(12):1572-3.
consensus_clustering_scExp( scExp, prefix = NULL, maxK = 10, reps = 100, pItem = 0.8, pFeature = 1, distance = "pearson", clusterAlg = "hc", innerLinkage = "ward.D", finalLinkage = "ward.D", plot_consclust = "pdf", plot_icl = "png" )
Arguments
| scExp | A SingleCellExperiment object containing 'PCA' in reducedDims. |
|---|---|
| prefix | character value for output directory. Directory is created only if plot_consclust is not NULL. This title can be an abosulte or relative path. |
| maxK | integer value. maximum cluster number to evaluate. (10) |
| reps | integer value. number of subsamples. (100) |
| pItem | numerical value. proportion of items to sample. (0.8) |
| pFeature | numerical value. proportion of features to sample. (1) |
| distance | character value. 'pearson': (1 - Pearson correlation), 'spearman' (1 - Spearman correlation), 'euclidean', 'binary', 'maximum', 'canberra', 'minkowski' or custom distance function. ('pearson') |
| clusterAlg | character value. cluster algorithm. 'hc' heirarchical (hclust), 'pam' for paritioning around medoids, 'km' for k-means upon data matrix, 'kmdist' ('hc') for k-means upon distance matrices (former km option), or a function that returns a clustering. ('hc') |
| innerLinkage | hierarchical linkage method for subsampling. ('ward.D') |
| finalLinkage | hierarchical linkage method for consensus matrix. ('ward.D') |
| plot_consclust | character value. NULL - print to screen, 'pdf', 'png', 'pngBMP' for bitmap png, helpful for large datasets. ('pdf') |
| plot_icl | same as above for item consensus plot. ('png') |
Value
Returns a SingleCellExperiment object containing consclust list, calculated cluster consensus and item consensus scores in metadata.
Details
This functions takes as input a SingleCellExperiment object that must have 'PCA' in reducedDims and outputs a SingleCellExperiment object containing consclust list calculated cluster consensus and item consensus scores in metadata.
References
ConsensusClusterPlus package by Wilkerson, M.D., Hayes, D.N. (2010). ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics, 2010 Jun 15;26(12):1572-3.
Examples
data("scExp") scExp_cf = correlation_and_hierarchical_clust_scExp(scExp) scExp_cf = consensus_clustering_scExp(scExp)#>#>#>#>#>#>#>#>#>#>