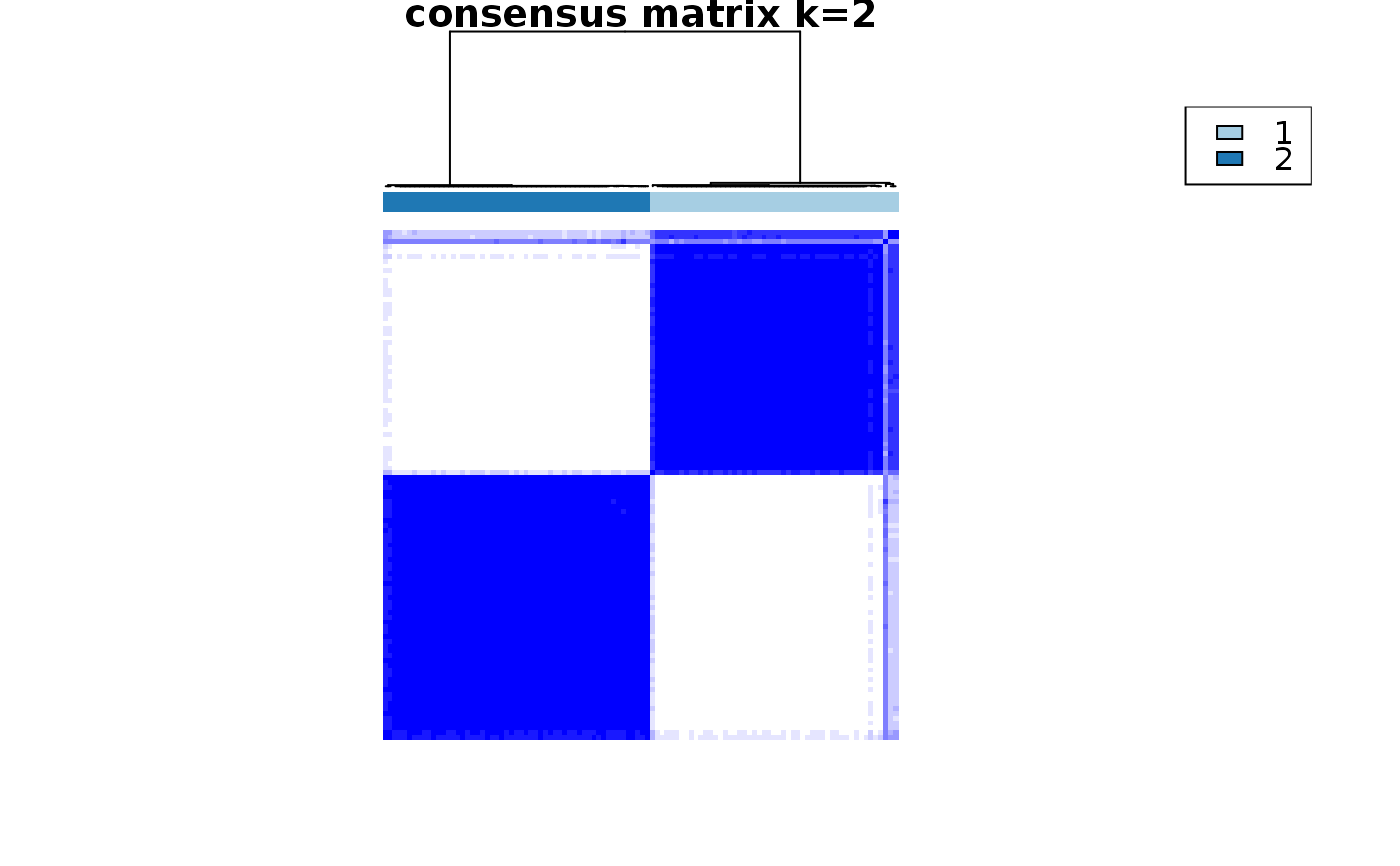
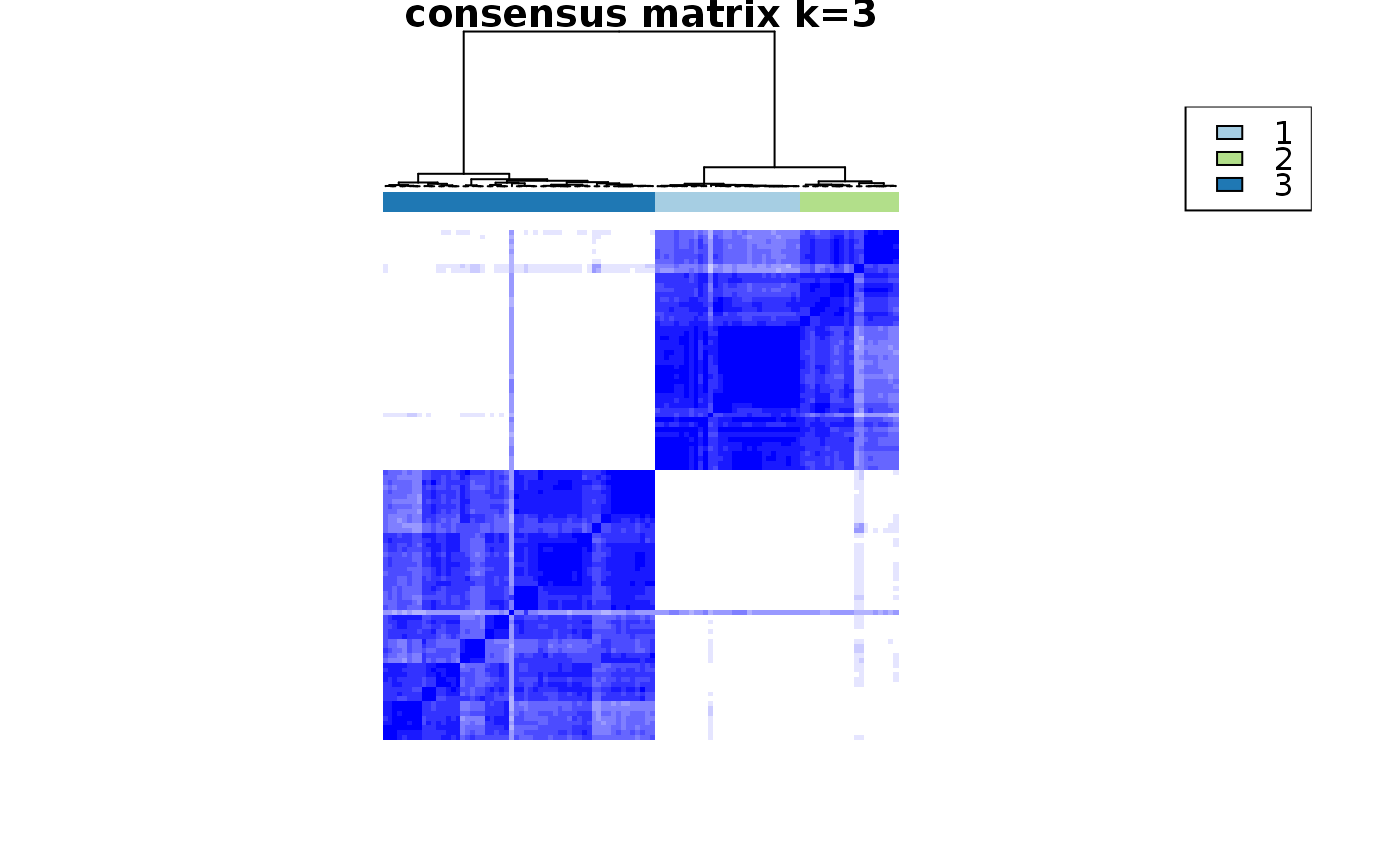
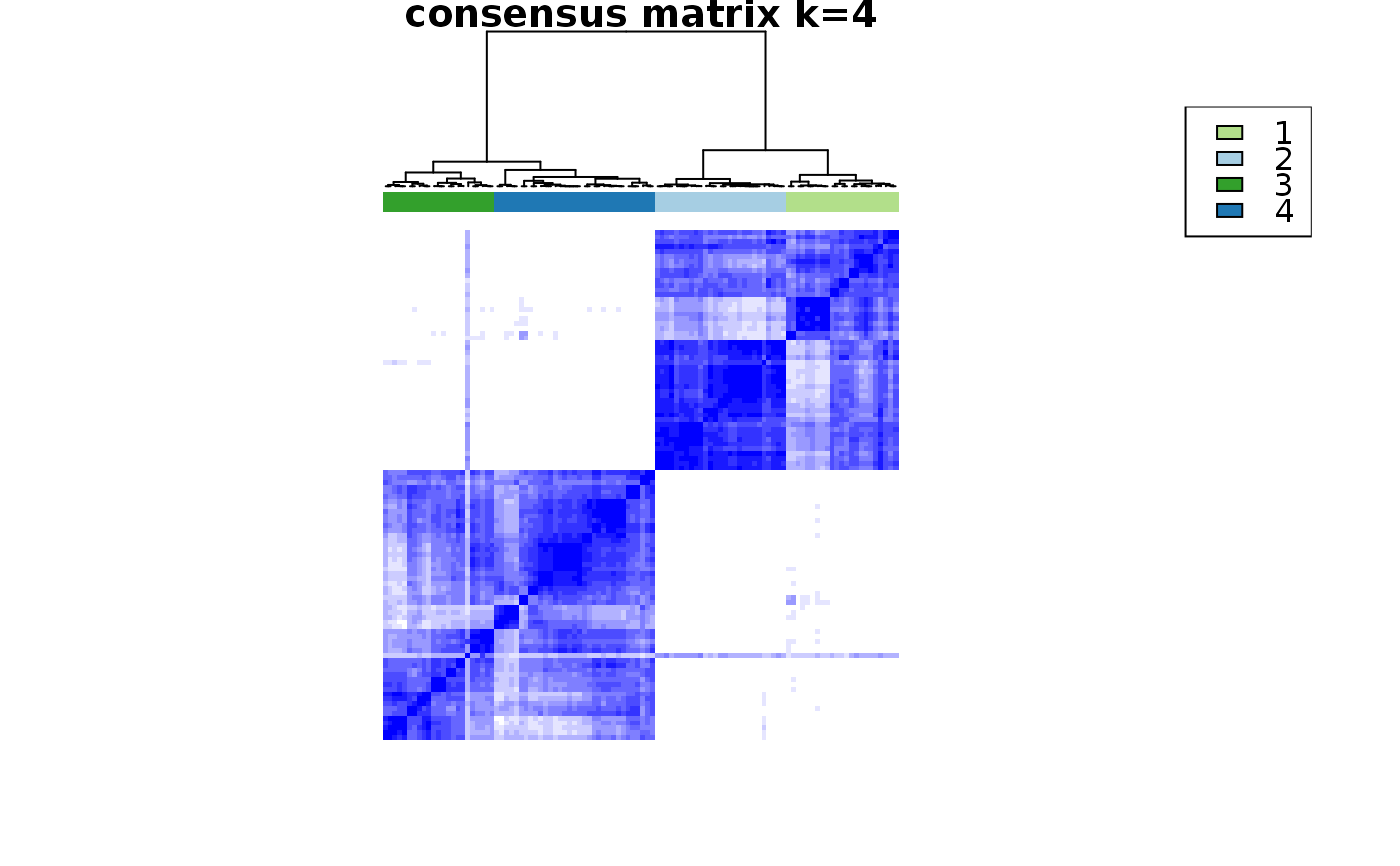
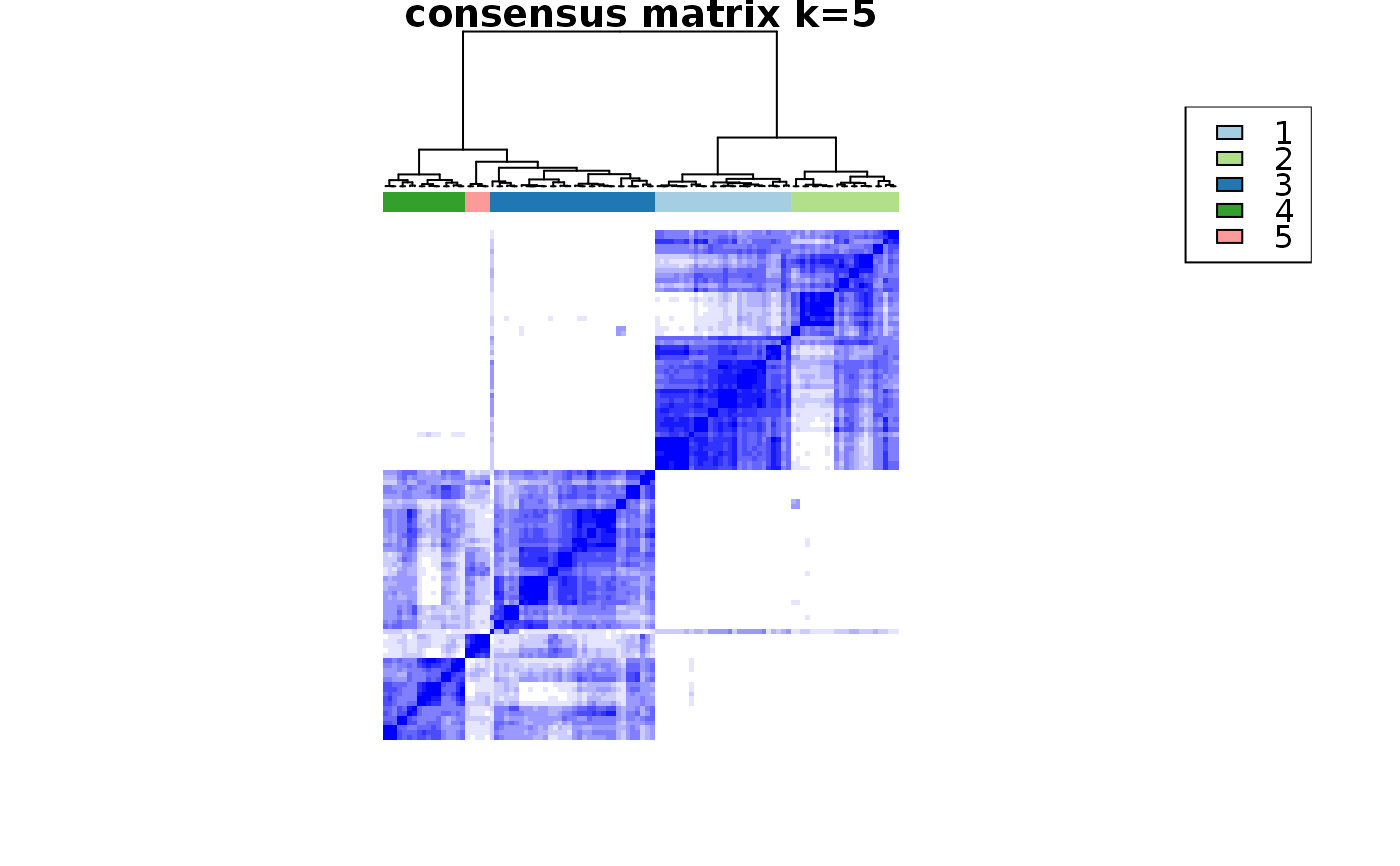
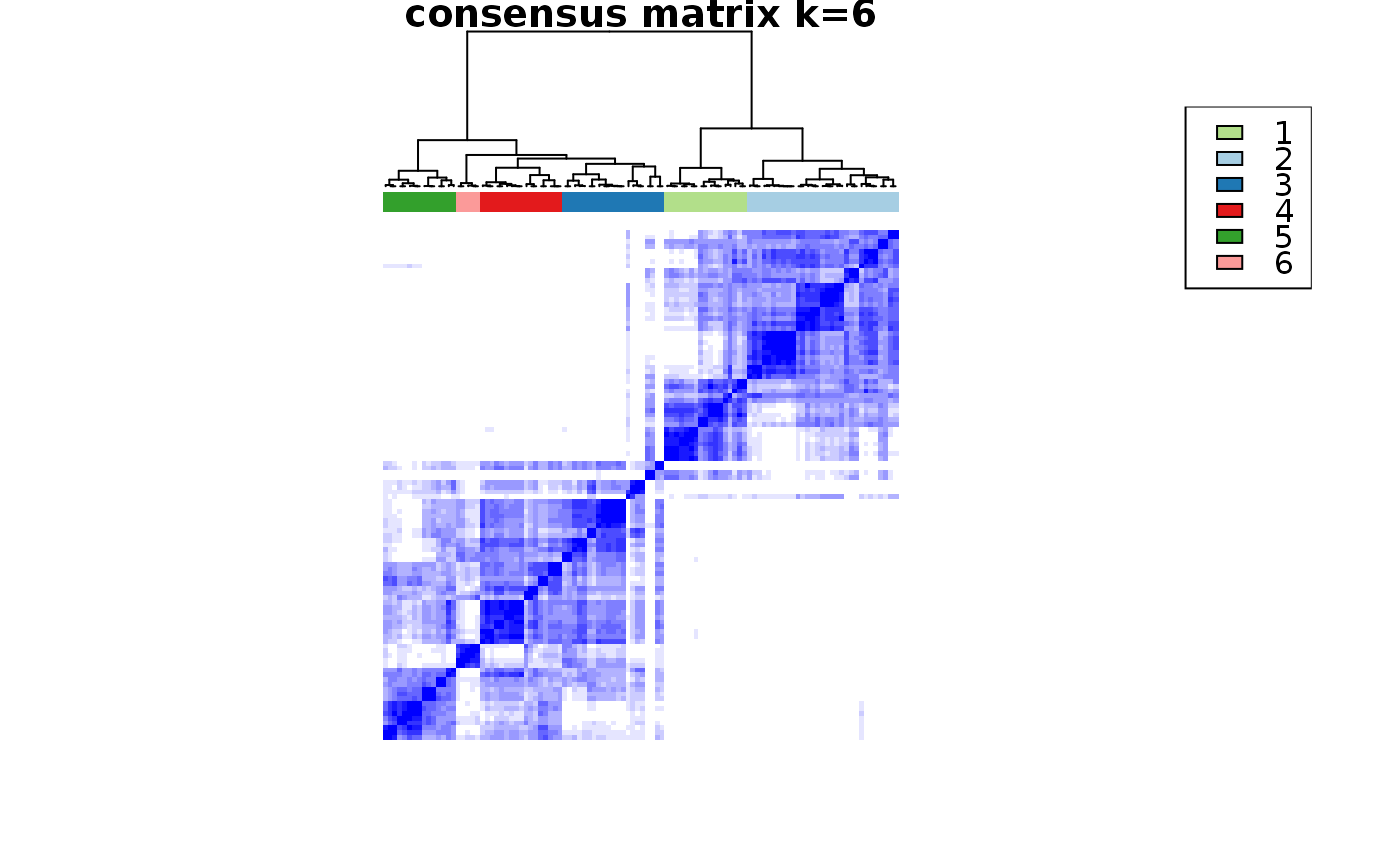
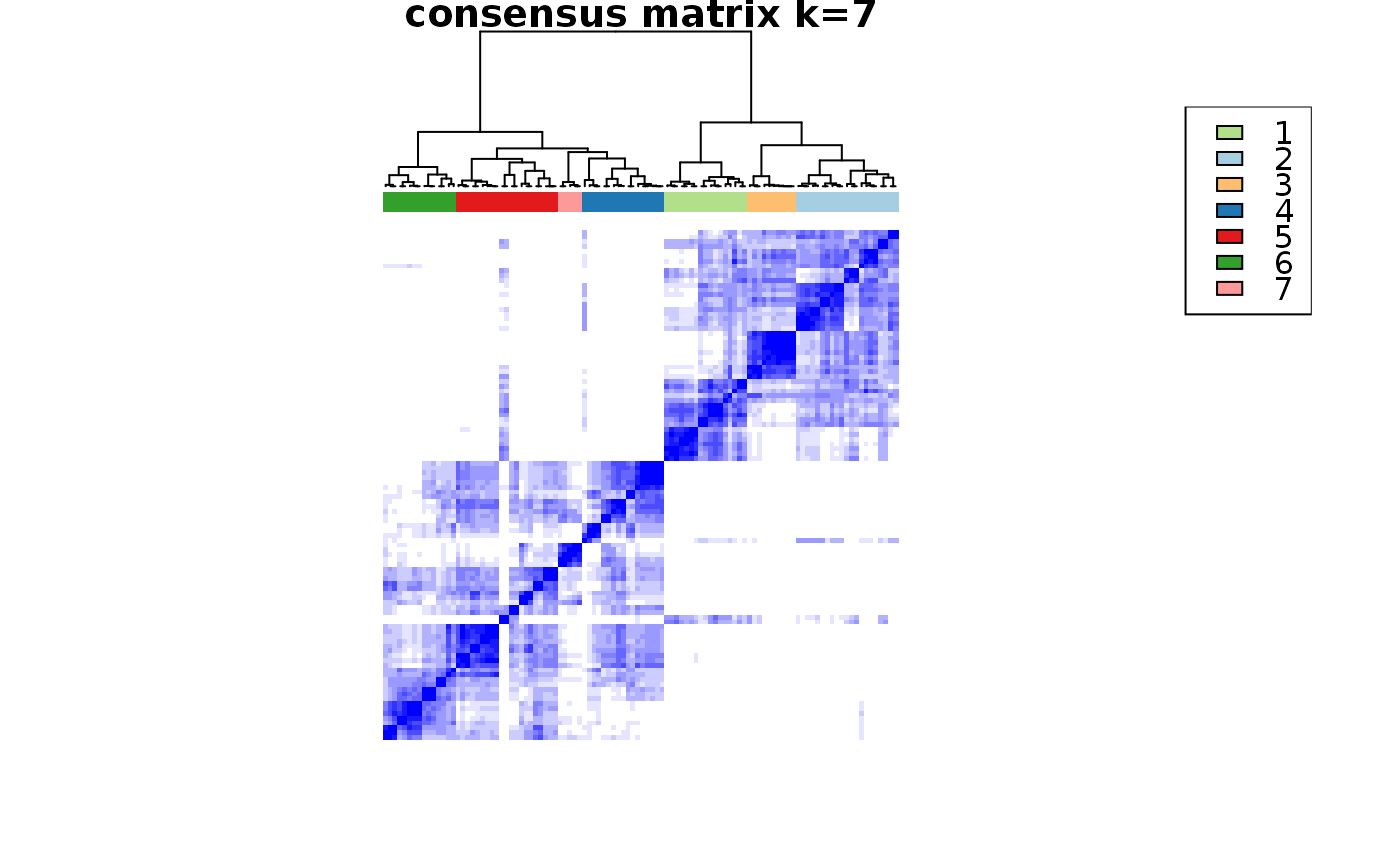
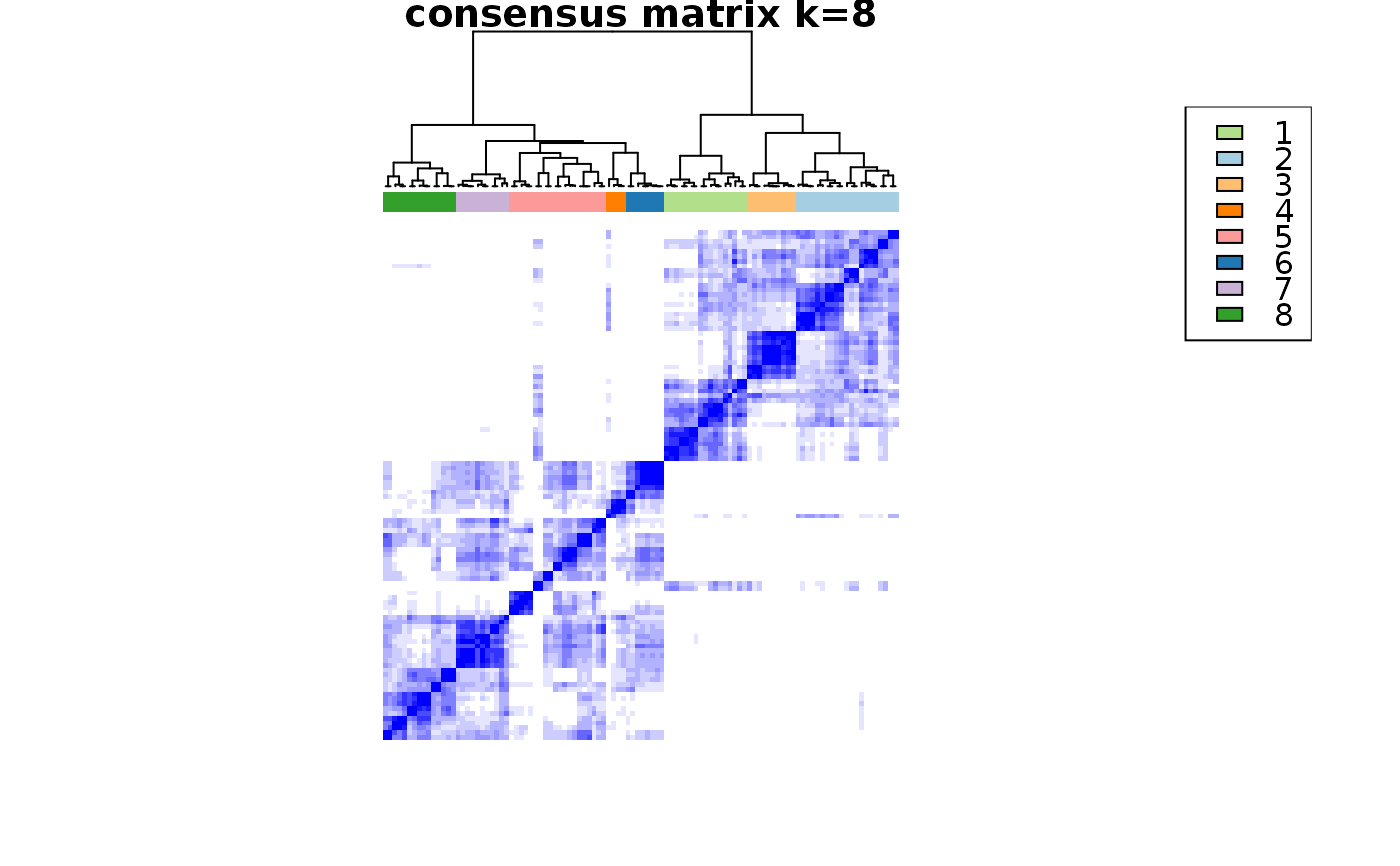
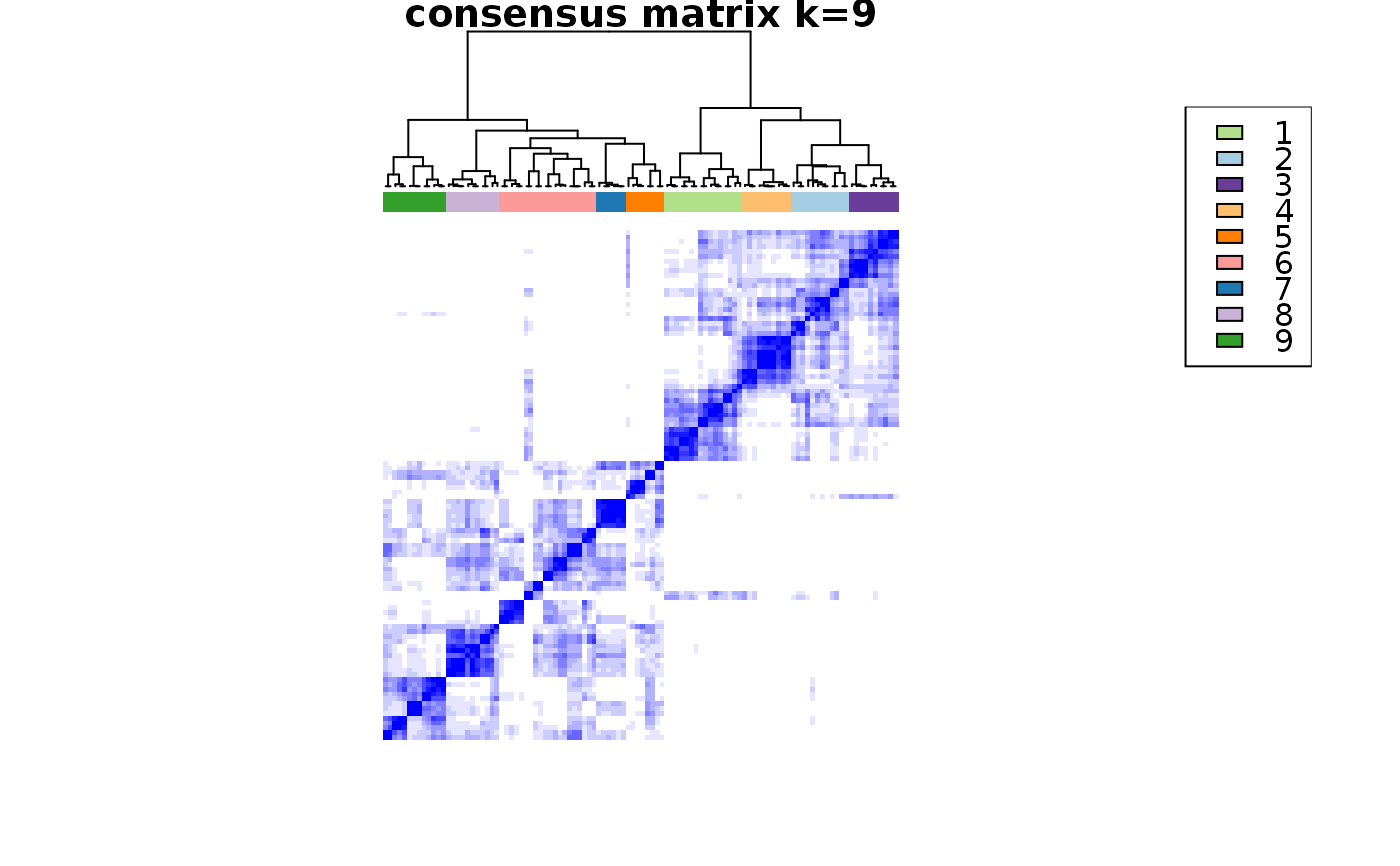
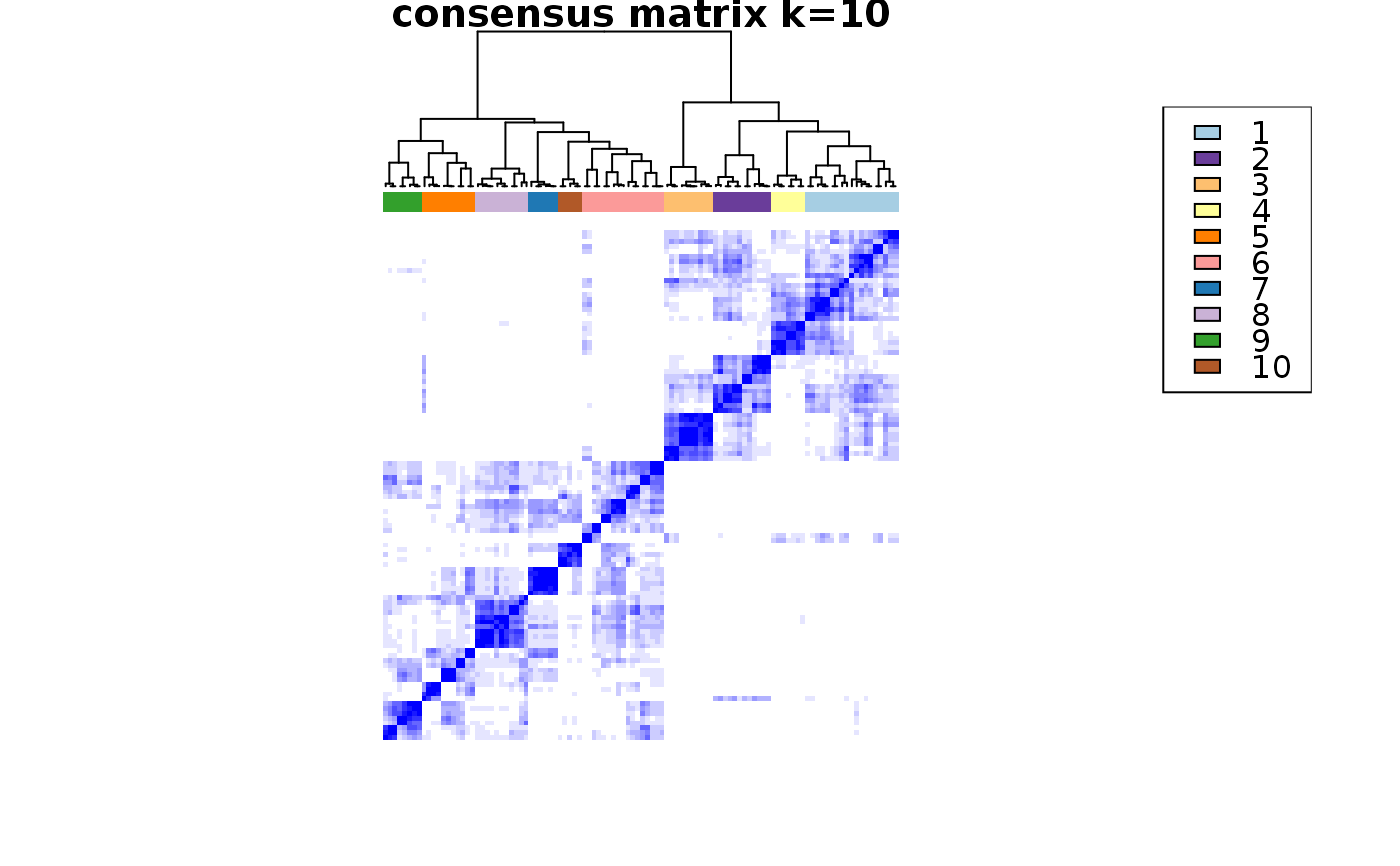
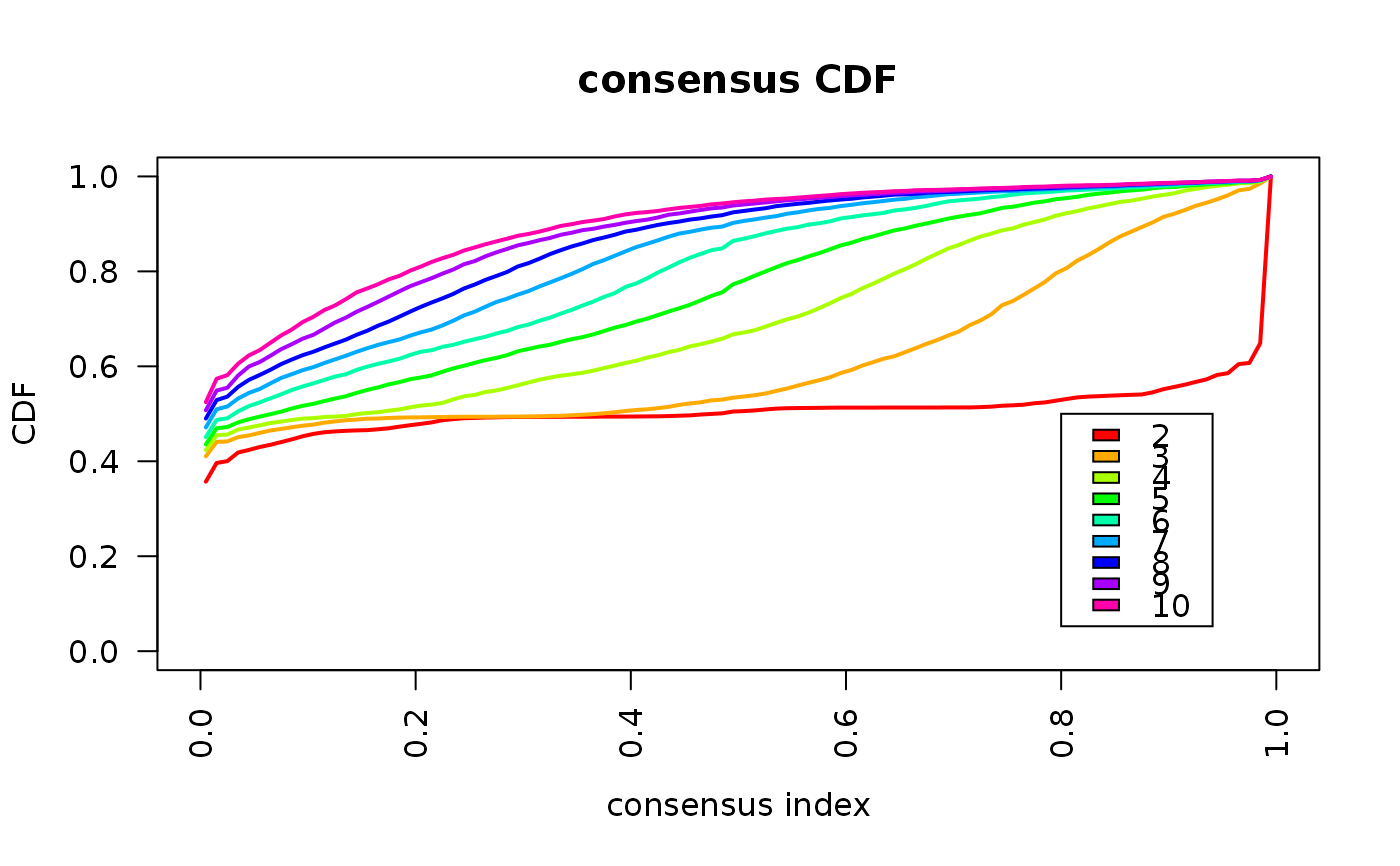
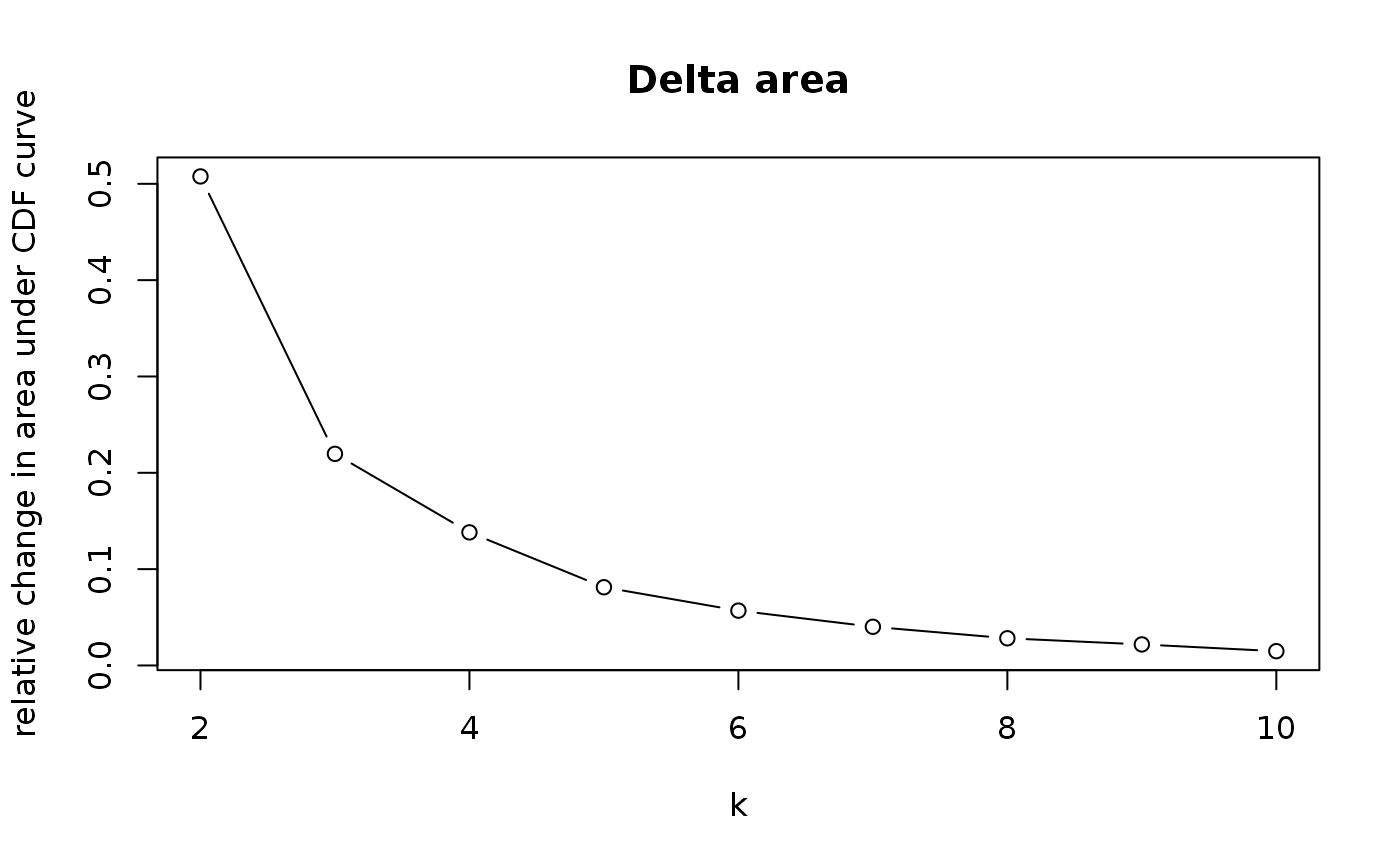
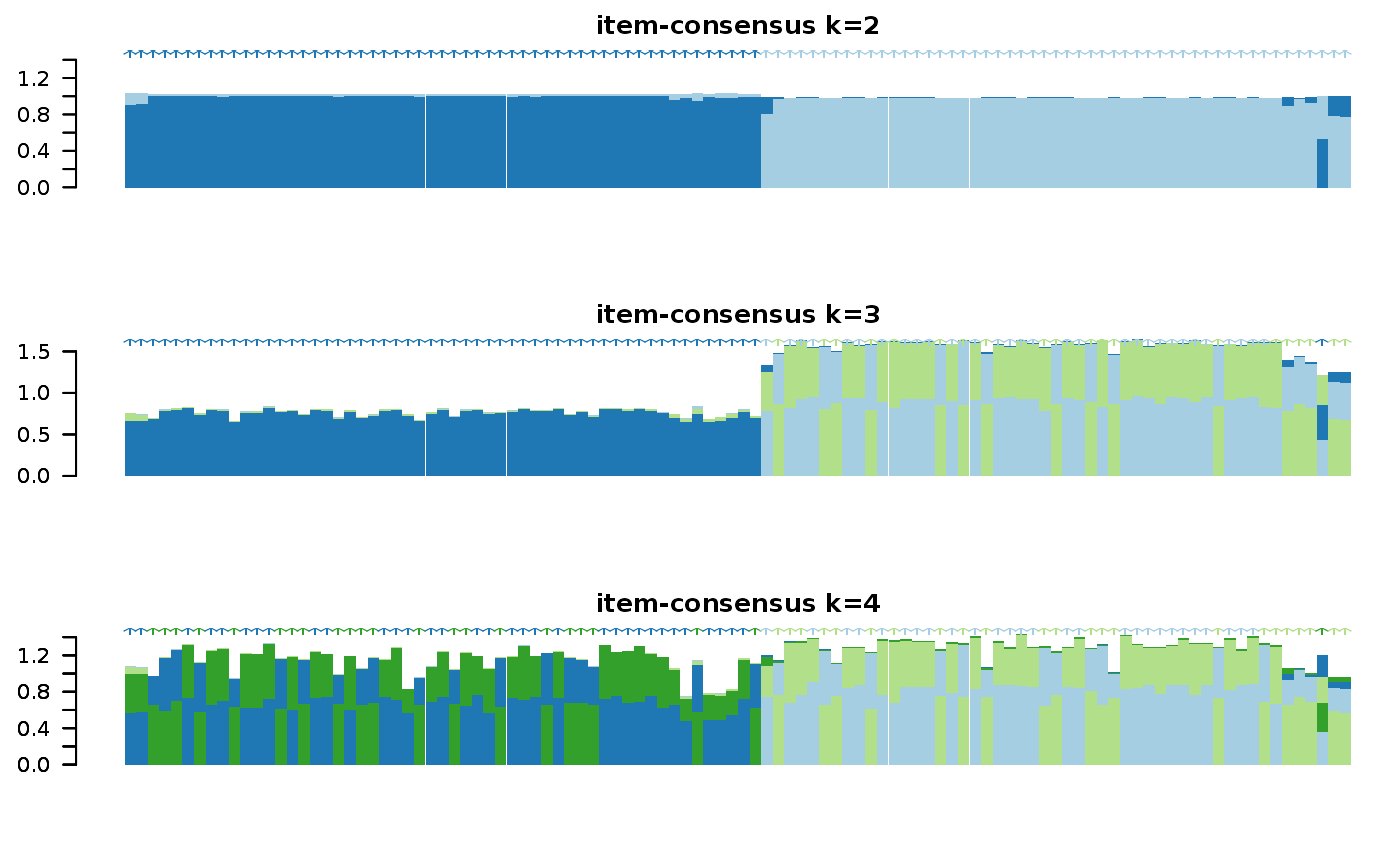
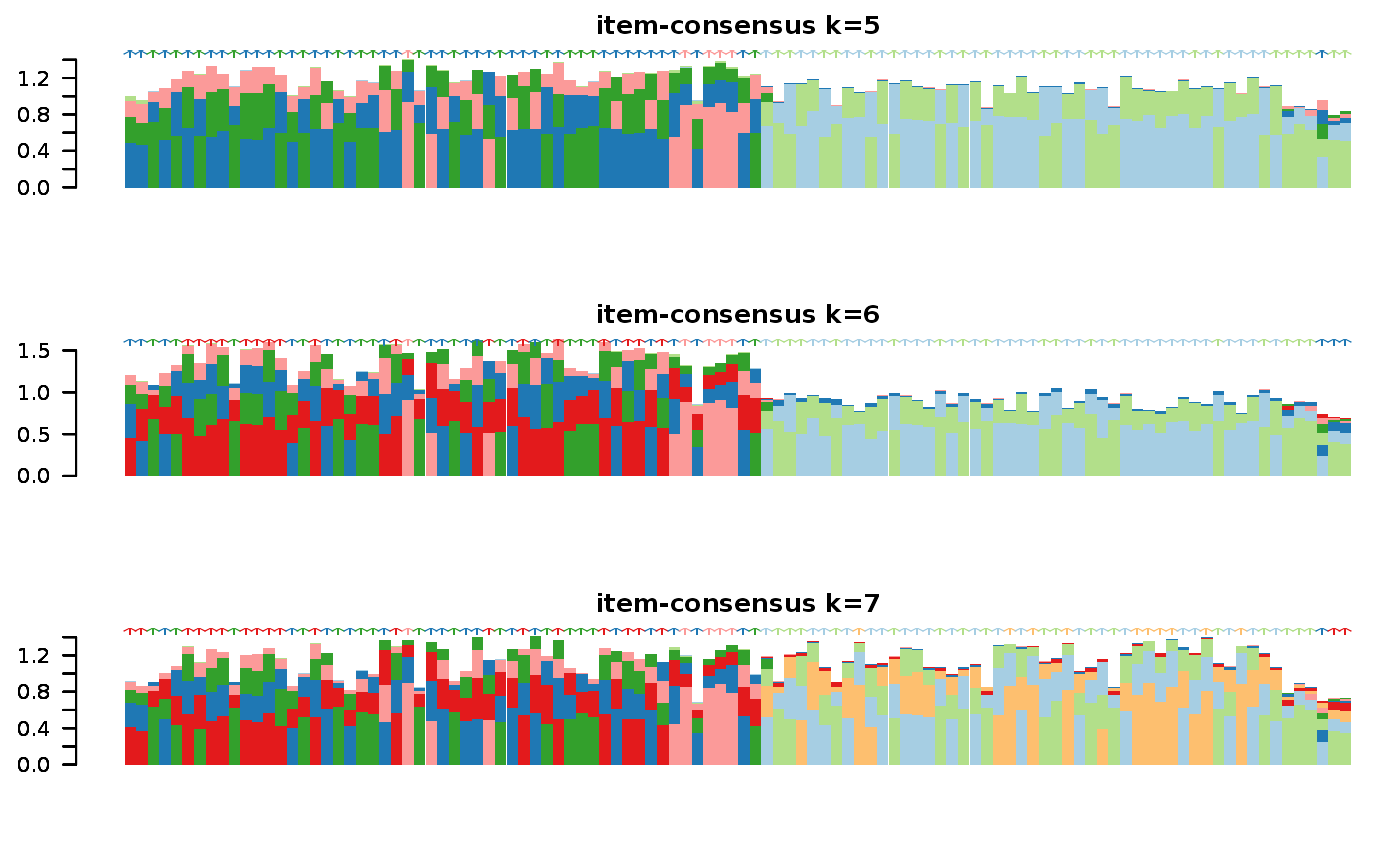
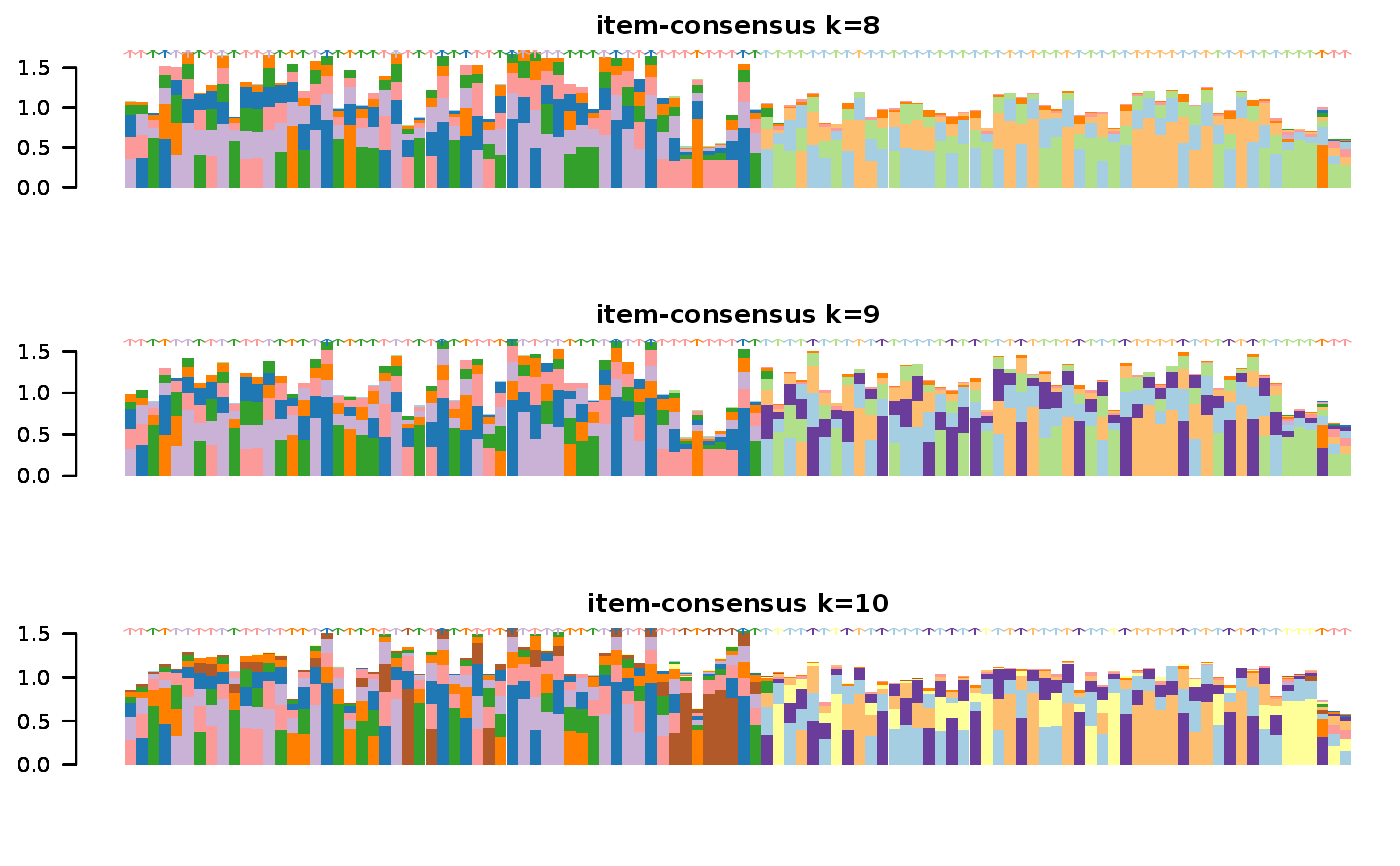
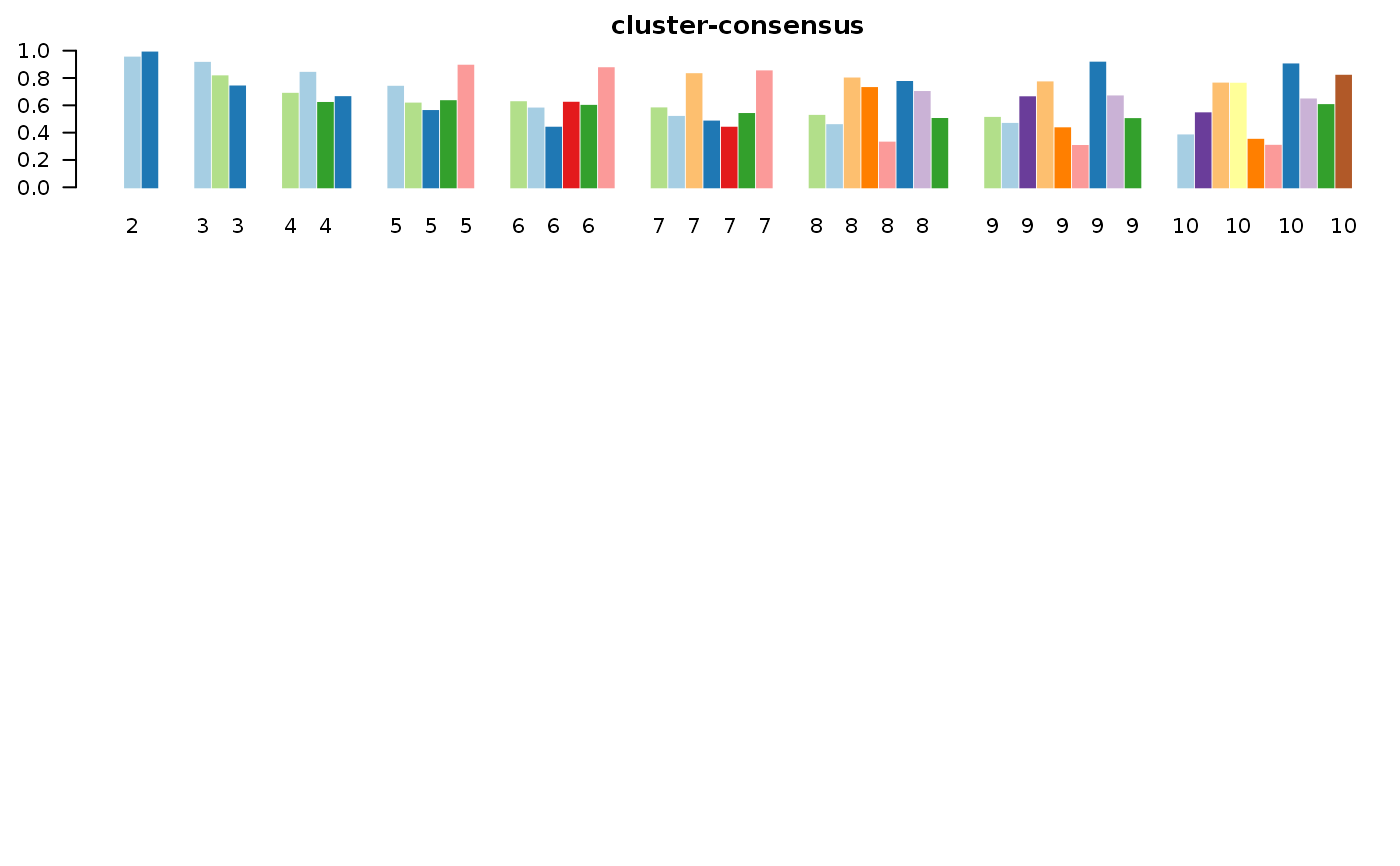This functions takes as input a SingleCellExperiment object with consclust and a number of cluster to select. It outputs a SingleCellExperiment object with each cell assigned to a correlation cluster in colData. Also calculates a hierarchical clustering of the consensus associations calculated by ConsensusClusterPlus.
choose_cluster_scExp( scExp, nclust = 3, consensus = FALSE, hc_linkage = "ward.D" )
Arguments
| scExp | A SingleCellExperiment object containing consclust in metadata. |
|---|---|
| nclust | Number of cluster to pick (3) |
| consensus | Use consensus clustering results instead of simple hierarchical clustering ? (FALSE) |
| hc_linkage | A linkage method for hierarchical clustering. See cor. ('ward.D') |
Value
Returns a SingleCellExperiment object with each cell assigned to a correlation cluster in colData.
Examples
data("scExp") scExp_cf = correlation_and_hierarchical_clust_scExp(scExp) scExp_cf = choose_cluster_scExp(scExp_cf,nclust=3,consensus=FALSE) table(scExp_cf$cell_cluster)#> #> C1 C2 C3 #> 47 40 19#>#>#>#>#>#>#>#>#>#>scExp_cf_consensus = choose_cluster_scExp(scExp_cf,nclust=3,consensus=TRUE) table(scExp_cf_consensus$cell_cluster)#> #> C1 C2 C3 #> 30 20 56